1 탐색적 데이터 분석
1.1 범주형 변수
범주형 변수(species, island, sex) 관계를 살펴보자.
1.1.1 species, island
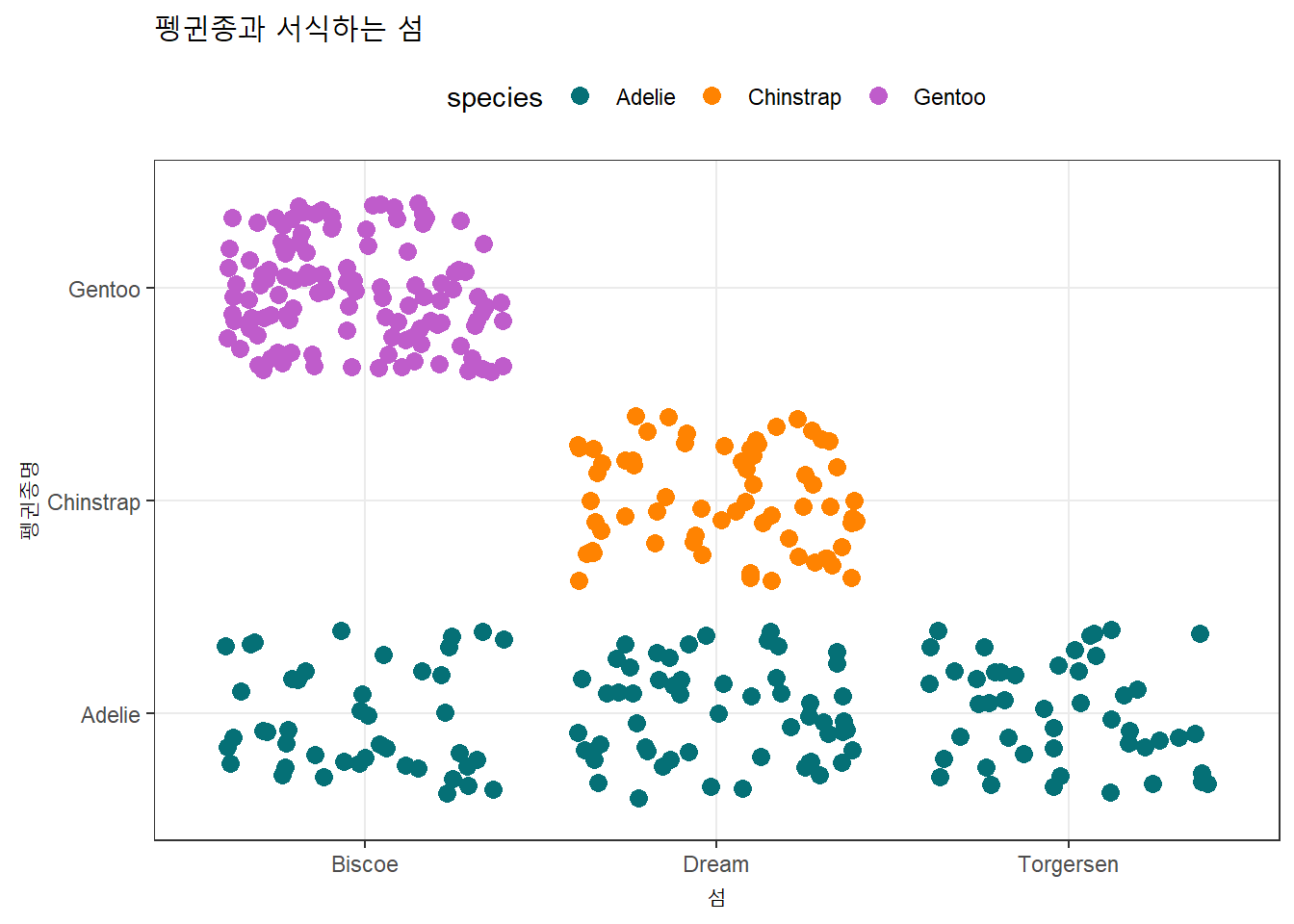
species, island 시각화, 표, 검정 통계량을 통해 서로 독립이 아님이 확인된다.
– Adelie: 아델리 펭귄은 모든 섬에 서식 – Gentoo: 젠투 펭귄은 비스코(Biscoe) 섬에만 서식 – Chinstrap: 턱끈 펭귄은 드림(Dream) 섬에만 서식.
시각화
코드
penguins_colors <- c('#057076', '#ff8301', '#bf5ccb')
penguins %>%
ggplot(aes(x = island, y = species, color = species)) +
geom_jitter(size = 3) +
scale_color_manual(values = penguins_colors) +
theme_bw(base_family = "NanumGothic") +
theme(legend.position = "top") +
labs(title = "펭귄종과 서식하는 섬",
x = "섬",
y = "펭귄종명") 표
코드
library(gtExtras)
penguins %>%
count(species, island) %>%
pivot_wider(names_from = island, values_from = n, values_fill = 0) %>%
gt::gt() %>%
gtExtras::gt_theme_538()| species | Biscoe | Dream | Torgersen |
|---|---|---|---|
| Adelie | 44 | 55 | 47 |
| Chinstrap | 0 | 68 | 0 |
| Gentoo | 119 | 0 | 0 |
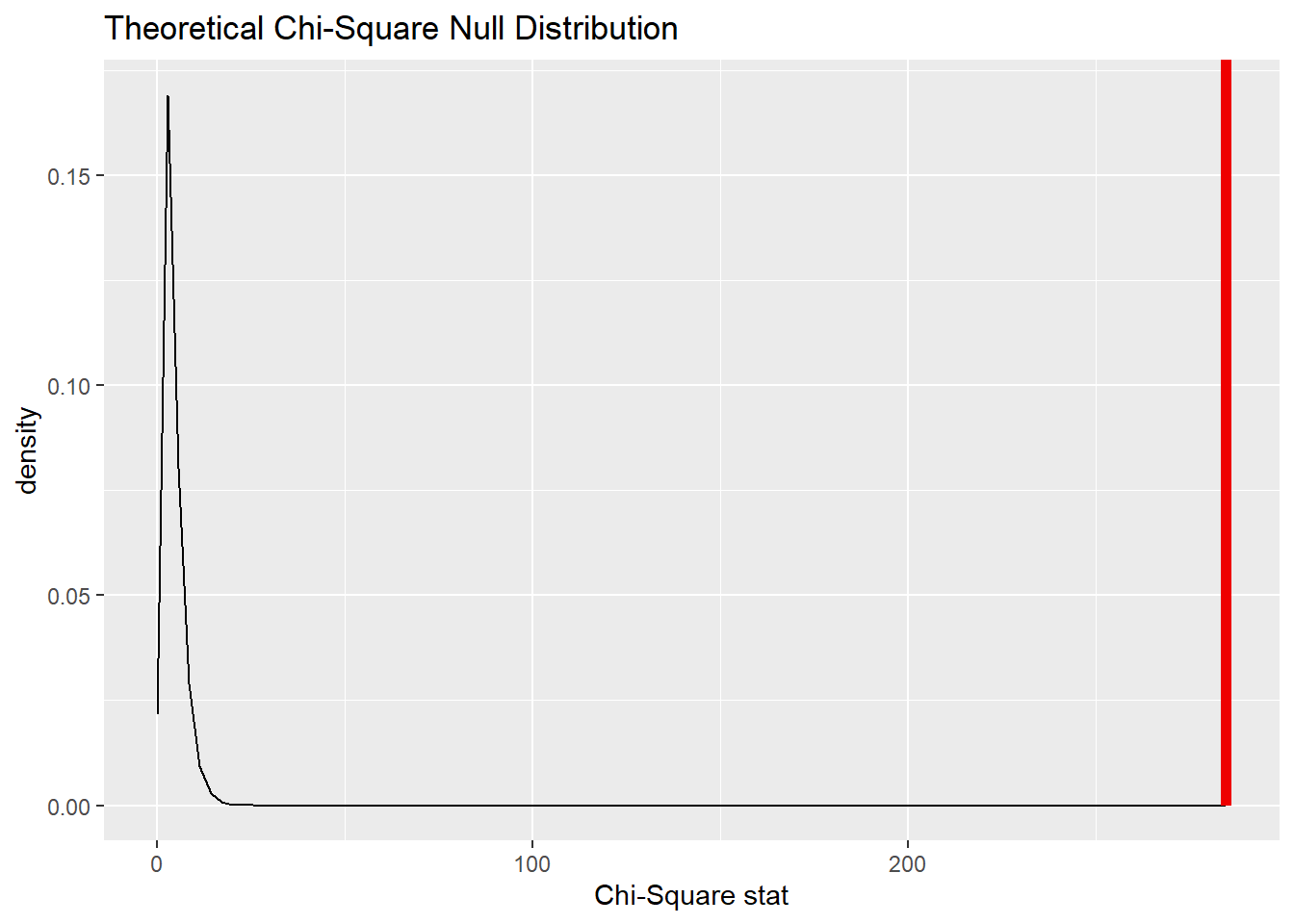
통계검정
코드
library(infer)
# 관측통계량
observed_indep_statistic <- penguins %>%
specify(species ~ island) %>%
hypothesize(null = "independence") %>%
calculate(stat = "Chisq")
# 통계검정 시각화
penguins %>%
specify(species ~ island) %>%
assume(distribution = "Chisq") %>%
visualize() +
shade_p_value(observed_indep_statistic,
direction = "greater") 다른 species와 sex, sex와 island 범주 사이는 특별한 관계를 찾을 수 없음.
species와 sex
코드
library(janitor)
library(gt)
penguins %>%
count(species, sex) %>%
pivot_wider(names_from = sex, values_from = n, values_fill = 0) %>%
adorn_totals("row", name = "합계") %>%
adorn_percentages("row") %>%
adorn_pct_formatting(digits = 1) %>%
adorn_ns() %>%
gt::gt() %>%
tab_options(table.font.names = 'NanumGothic',
column_labels.font.weight = 'bold',
heading.title.font.size = 14,
heading.subtitle.font.size = 14,
table.font.color = 'steelblue',
source_notes.font.size = 10,
#source_notes.
table.font.size = 14) %>%
gtExtras::gt_theme_538()| species | female | male |
|---|---|---|
| Adelie | 50.0% (73) | 50.0% (73) |
| Chinstrap | 50.0% (34) | 50.0% (34) |
| Gentoo | 48.7% (58) | 51.3% (61) |
| 합계 | 49.5% (165) | 50.5% (168) |
sex와 island
코드
penguins %>%
count(island, sex) %>%
pivot_wider(names_from = sex, values_from = n, values_fill = 0) %>%
adorn_totals("row", name = "합계") %>%
adorn_percentages("row") %>%
adorn_pct_formatting(digits = 1) %>%
adorn_ns() %>%
gt::gt() %>%
tab_options(table.font.names = 'NanumGothic',
column_labels.font.weight = 'bold',
heading.title.font.size = 14,
heading.subtitle.font.size = 14,
table.font.color = 'darkgreen',
source_notes.font.size = 10,
#source_notes.
table.font.size = 14) %>%
gtExtras::gt_theme_538()| island | female | male |
|---|---|---|
| Biscoe | 49.1% (80) | 50.9% (83) |
| Dream | 49.6% (61) | 50.4% (62) |
| Torgersen | 51.1% (24) | 48.9% (23) |
| 합계 | 49.5% (165) | 50.5% (168) |
1.2 연속형 변수
1.2.1 상관관계
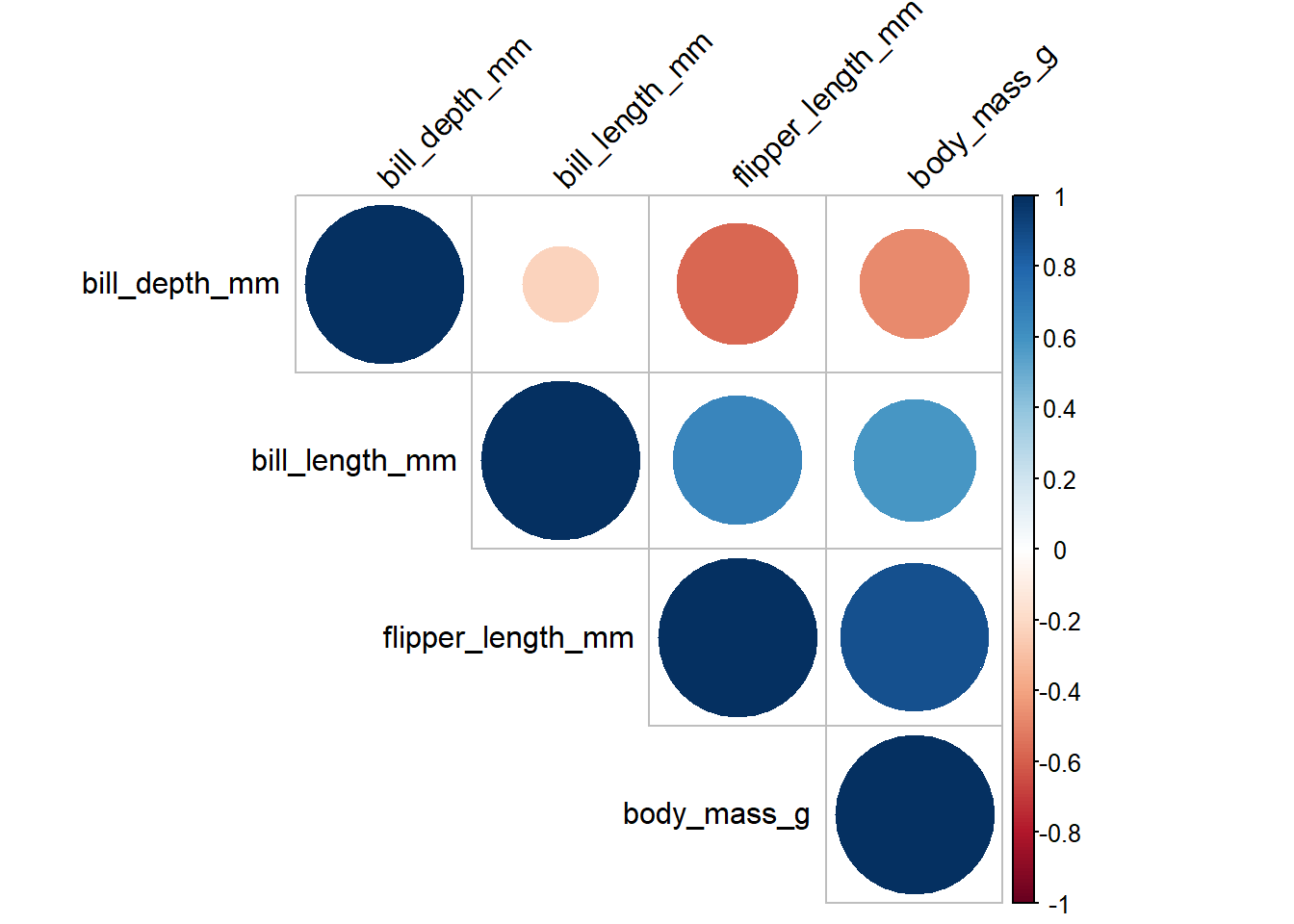
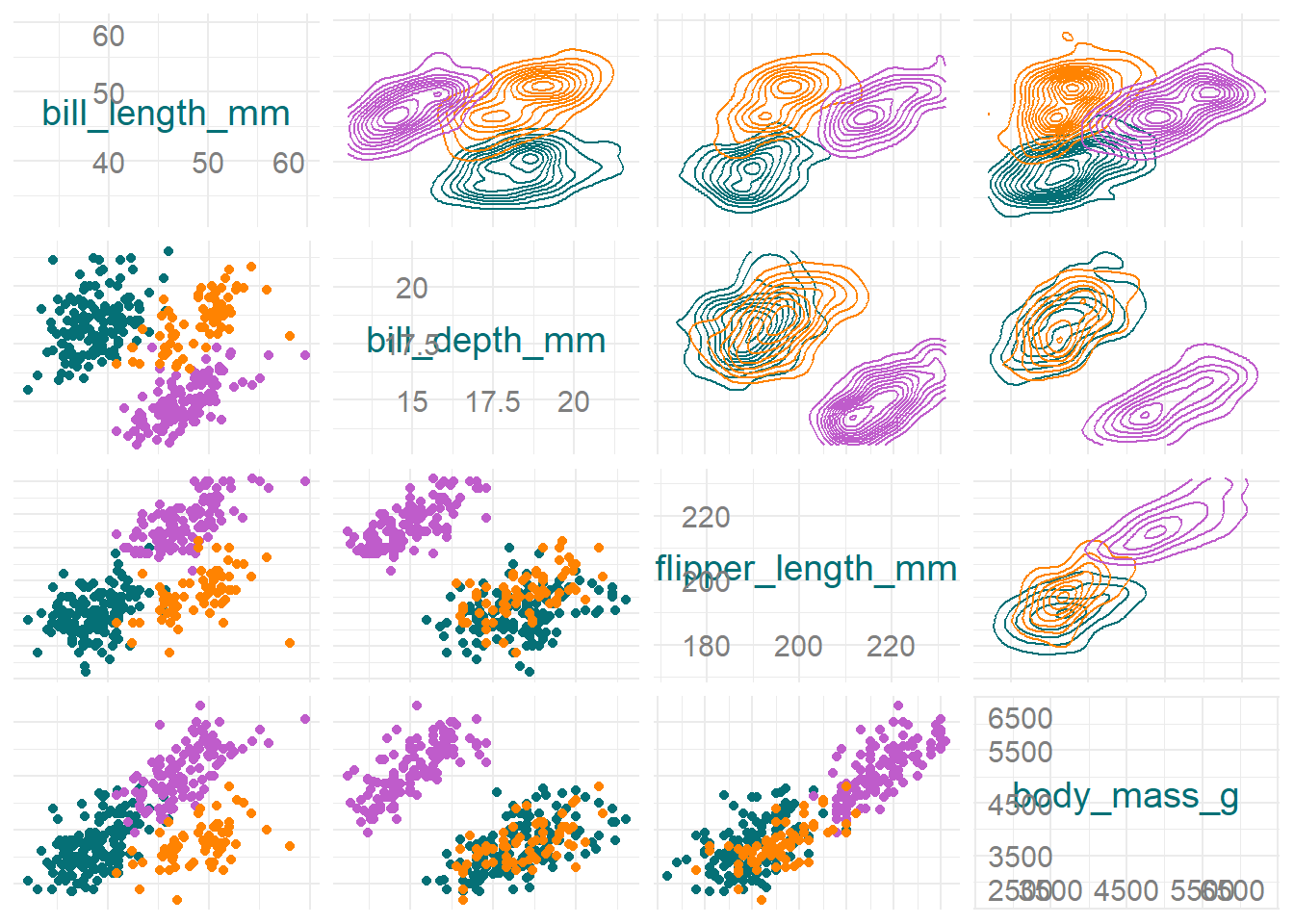
penguins 데이터프레임에서 연속형 변수만 추출하여 상관관계를 살펴보자.
시각화
코드
상관계수 일반표
코드
library(corrr)
library(gt)
penguins_num %>%
rename_with(.cols = everything(), .fn = ~str_remove(., pattern = "_mm|_g")) %>%
corrr::correlate( use = "pairwise.complete.obs",
method = "pearson") %>%
rearrange() %>%
shave() %>%
# fashion() %>%
gt::gt() %>%
# 상관계수 색상
data_color(
columns = where(is.numeric),
colors = scales::col_numeric(
## option D for Viridis - correlation coloring
palette = RColorBrewer::brewer.pal(9, "RdBu"),
domain = NULL,
na.color = 'white')
) %>%
sub_missing(
columns = everything(),
missing_text = "-"
) %>%
cols_align(align = "center", columns = everything()) %>%
gtExtras::gt_theme_538() | term | flipper_length | body_mass | bill_length | bill_depth |
|---|---|---|---|---|
| flipper_length | - | - | - | - |
| body_mass | 0.8729789 | - | - | - |
| bill_length | 0.6530956 | 0.5894511 | - | - |
| bill_depth | -0.5777917 | -0.4720157 | -0.2286256 | - |
상관계수 긴 표
코드
corrr::correlate(penguins_num) %>%
stretch(remove.dups = TRUE) %>%
filter(!is.na(r)) %>%
arrange(desc(r)) %>%
mutate(음양 = ifelse(r > 0, "양의 상관", "음의 상관")) %>%
mutate(color = "") %>%
gt(groupname_col = "음양") %>%
fmt_number(
columns = r,
decimals = 2,
use_seps = FALSE
) %>%
data_color(
columns = r,
target_columns = color,
method = "numeric",
palette = RColorBrewer::brewer.pal(9, "RdBu")
) %>%
cols_label(
x = "",
y = "",
r = "상관계수",
color = ""
) |>
opt_vertical_padding(scale = 0.7) %>%
gtExtras::gt_theme_538()| 상관계수 | |||
|---|---|---|---|
| 양의 상관 | |||
| flipper_length_mm | body_mass_g | 0.87 | |
| bill_length_mm | flipper_length_mm | 0.65 | |
| bill_length_mm | body_mass_g | 0.59 | |
| 음의 상관 | |||
| bill_length_mm | bill_depth_mm | −0.23 | |
| bill_depth_mm | body_mass_g | −0.47 | |
| bill_depth_mm | flipper_length_mm | −0.58 | |
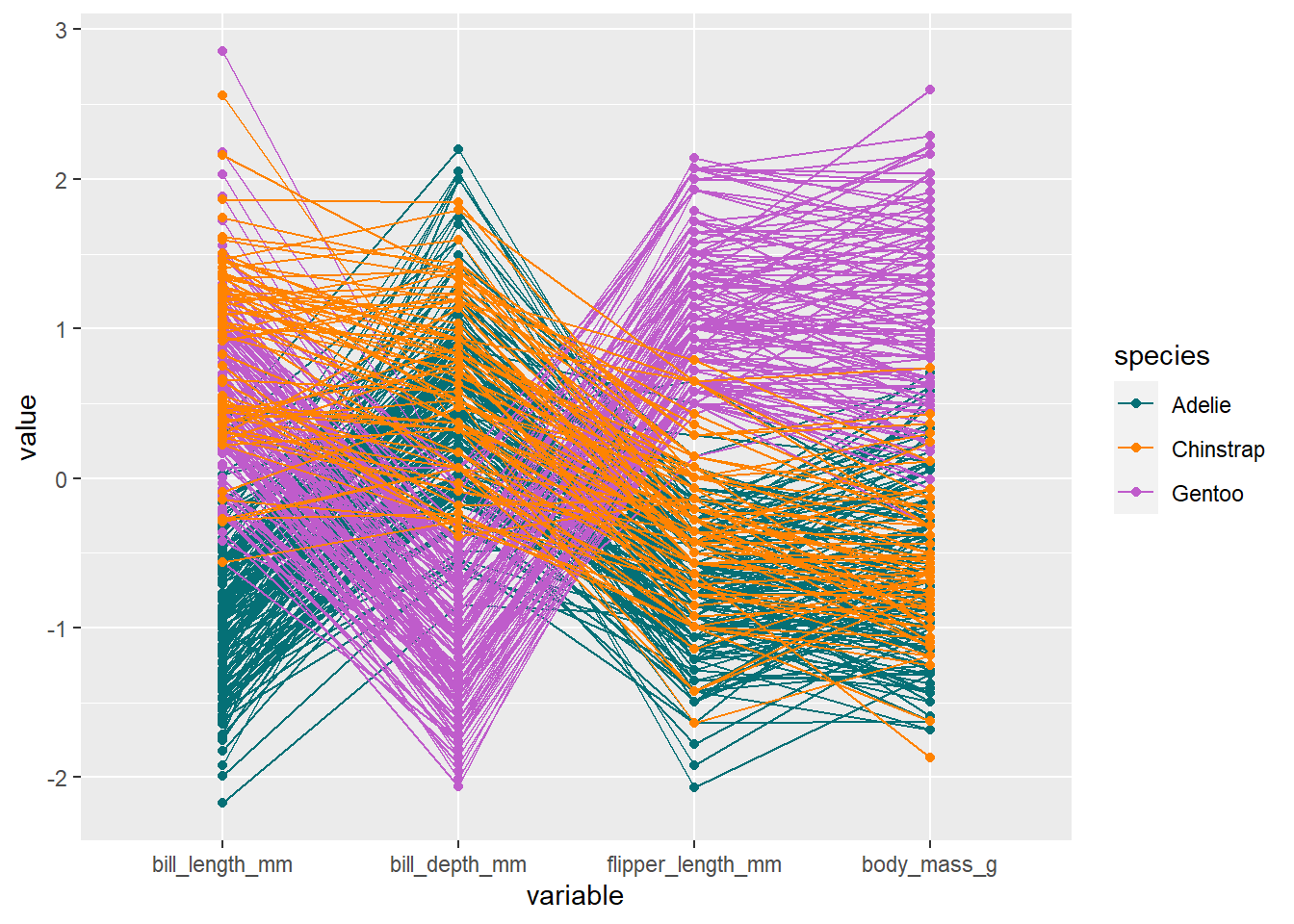
1.2.2 평행 그래프
코드
library(GGally)
penguins %>%
select(species, bill_length_mm, bill_depth_mm, flipper_length_mm, body_mass_g) %>%
ggparcoord(columns = 2:5,
showPoints = TRUE,
groupColumn = "species") +
scale_color_manual(values = penguins_colors)1.2.3 짝 산점도
코드
penguins %>%
select(species, bill_length_mm, bill_depth_mm, flipper_length_mm, body_mass_g) %>%
ggpairs(columns = 2:5, ggplot2::aes(colour=species),
upper = list(continuous='density', combo='box_no_facet'),
axisLabels='internal') +
scale_color_manual(values = penguins_colors) +
scale_fill_manual(values = penguins_colors) +
theme_minimal()2 모형
2.1 성별 분류모형
펭귄의 암수를 분류하는 기계학습모형을 개발하기 전에 훈련/시험 데이터셋으로 나눈다.
코드
library(tidymodels)
penguins_split <- penguins %>%
initial_split(prop = 0.70, strata = sex)
penguins_train <- training(penguins_split)
penguins_test <- testing(penguins_split)2.1.1 로지스틱 회귀
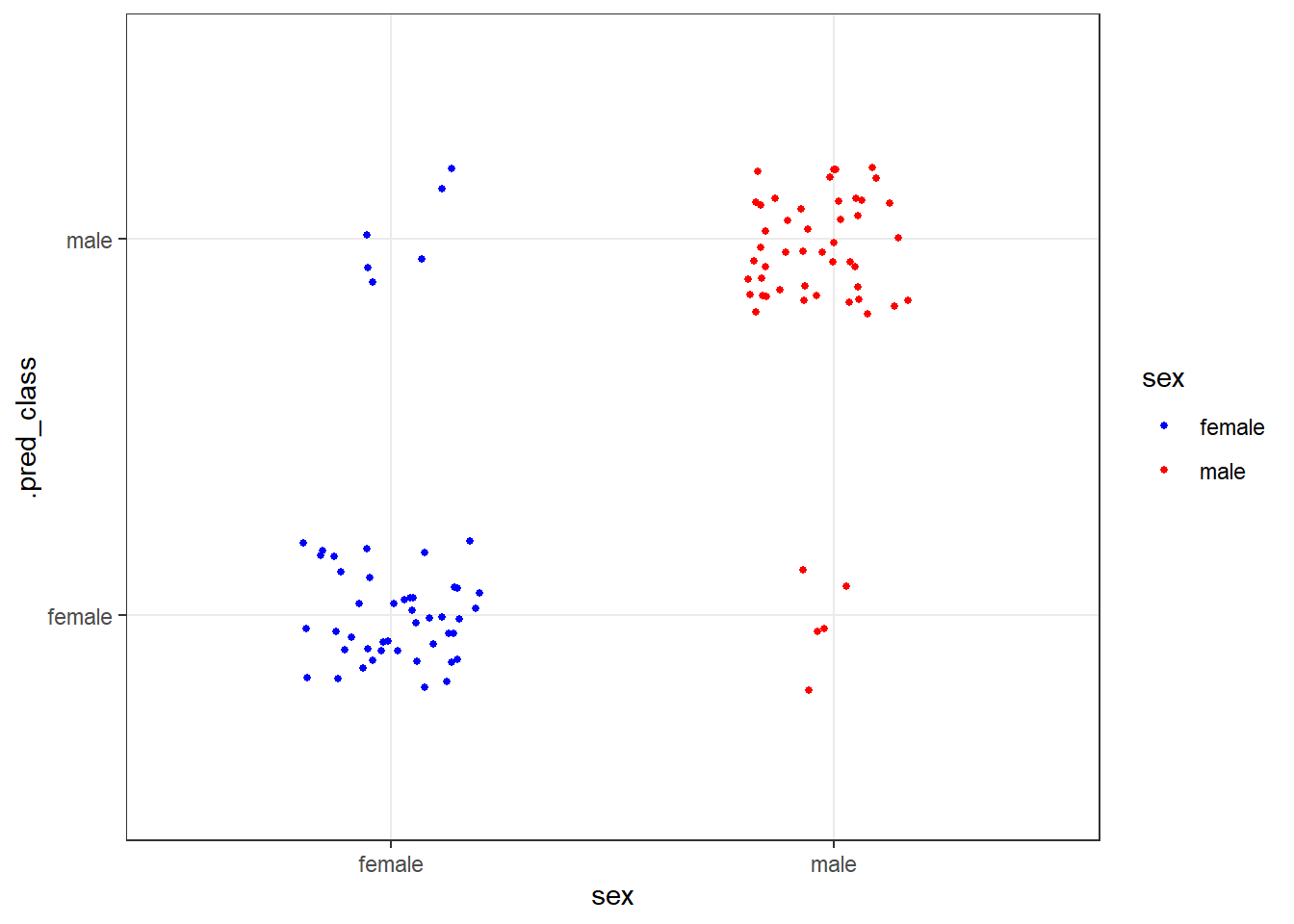
펭귄의 암수를 분류하는 로지스틱 회귀모형으로 분류모형을 개발해보자.
코드
logistic_spec <- logistic_reg() %>%
set_engine("glm") %>%
set_mode("classification")
logistic_wflow <-
workflow(
sex ~ species + island + bill_length_mm + bill_depth_mm +
flipper_length_mm + body_mass_g,
logistic_spec
)
logistic_fit <- logistic_wflow %>% fit(data = penguins_train)
sex_predict <- predict(logistic_fit, penguins_test, type = "class")
bind_cols(penguins_test, sex_predict) %>%
ggplot(aes(x = sex, y = .pred_class, color = sex)) +
geom_jitter(size = 1, width = 0.2, height = 0.2) +
scale_color_manual(values = c("blue", "red")) +
theme_bw()2.1.2 Random Forest
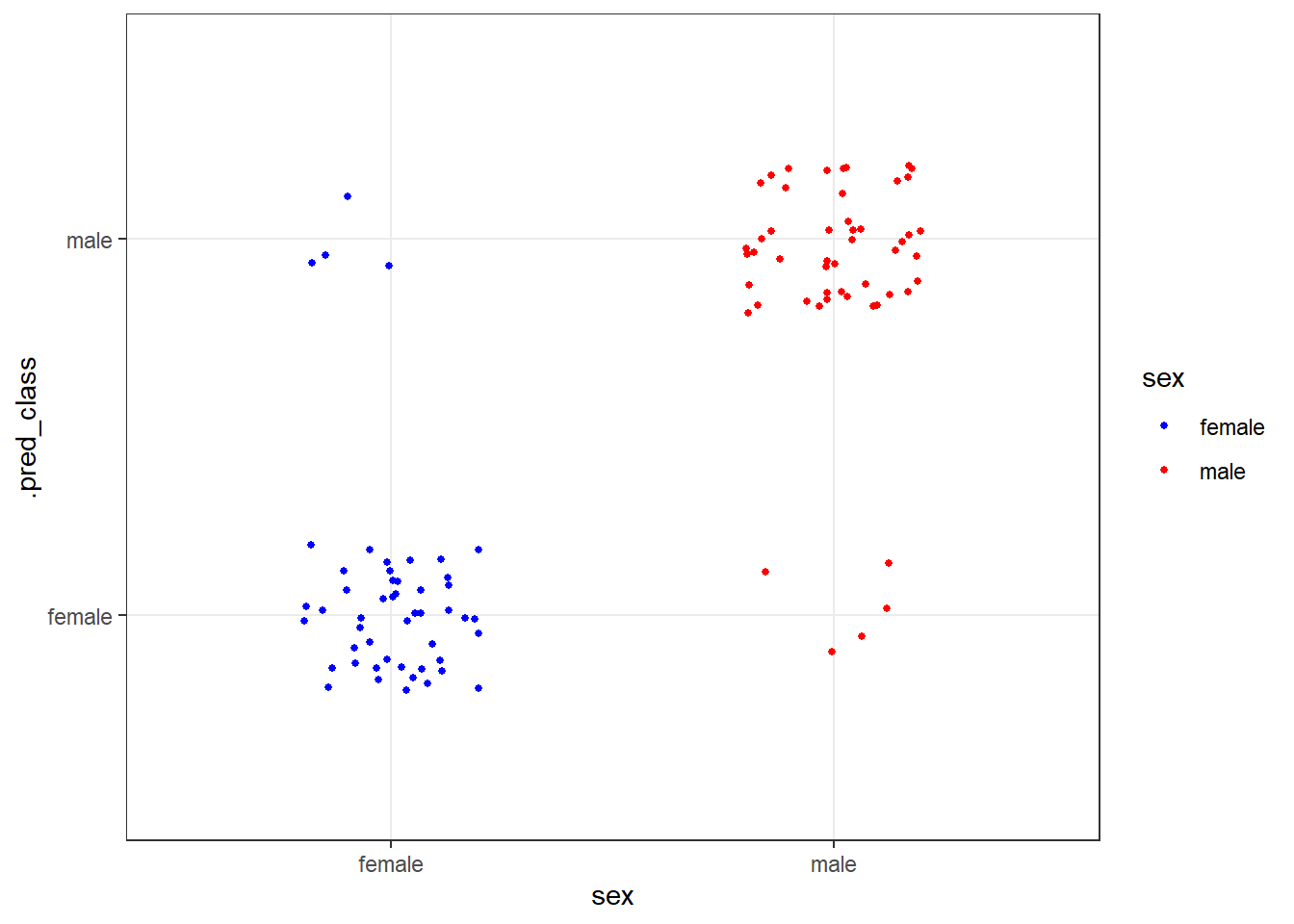
펭귄의 암수를 분류하는 Random Forest 모형으로 분류모형을 개발해보자.
코드
rf_spec <- rand_forest() %>%
set_engine("ranger") %>%
set_mode("classification")
rf_wflow <-
workflow(
sex ~ species + island + bill_length_mm + bill_depth_mm +
flipper_length_mm + body_mass_g,
rf_spec
)
rf_fit <- rf_wflow %>% fit(data = penguins_train)
sex_rf <- predict(rf_fit, penguins_test, type = "class")
bind_cols(penguins_test, sex_rf) %>%
ggplot(aes(x = sex, y = .pred_class, color = sex)) +
geom_jitter(size = 1, width = 0.2, height = 0.2) +
scale_color_manual(values = c("blue", "red")) +
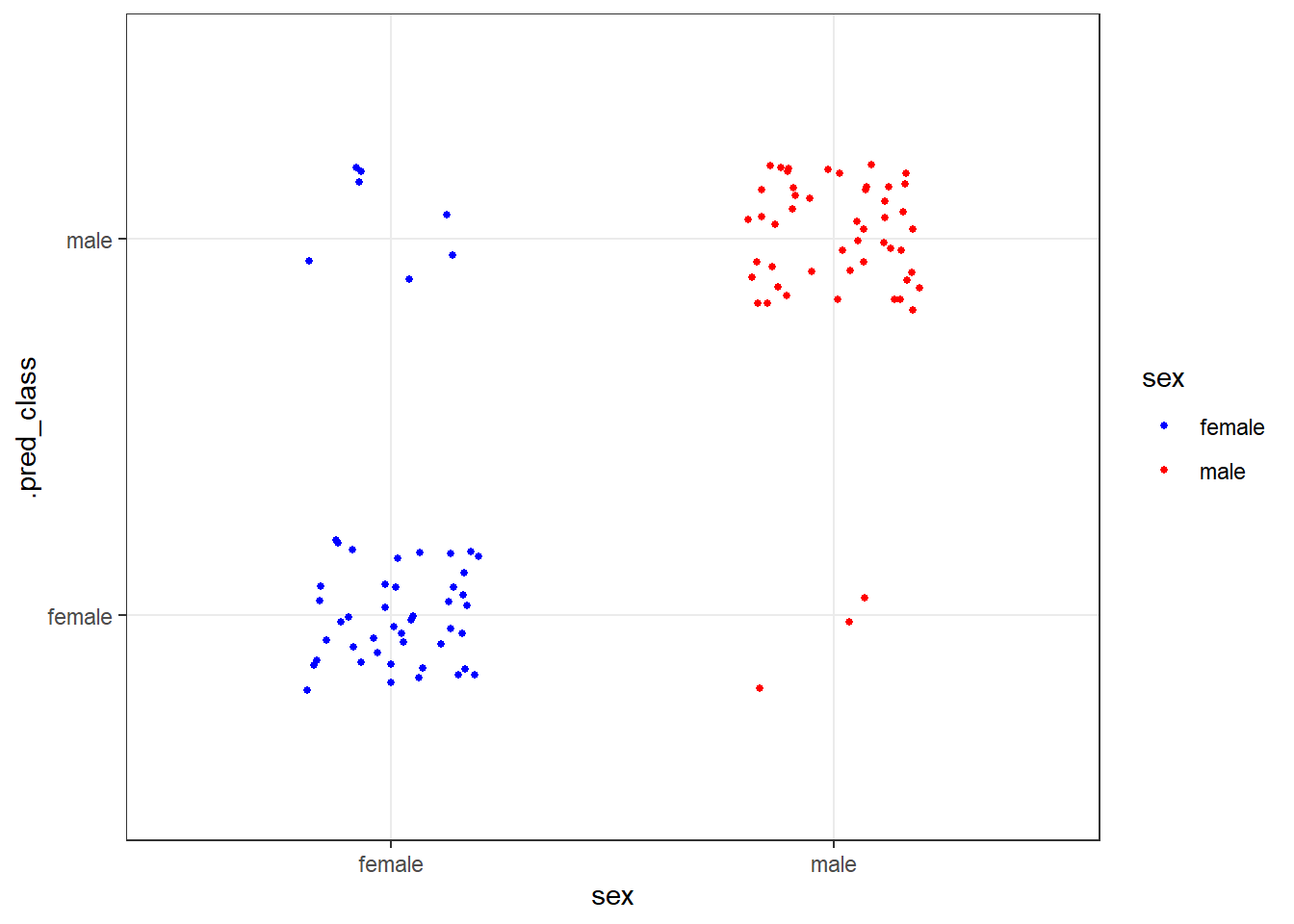
theme_bw()2.1.3 SVM
펭귄의 암수를 분류하는 SVM 모형으로 분류모형을 개발해보자.
코드
svm_spec <- svm_rbf() %>%
set_engine("kernlab") %>%
set_mode("classification")
svm_wflow <-
workflow(
sex ~ species + island + bill_length_mm + bill_depth_mm +
flipper_length_mm + body_mass_g,
svm_spec
)
svm_fit <- svm_wflow %>% fit(data = penguins_train)
sex_svm <- predict(svm_fit, penguins_test, type = "class")
bind_cols(penguins_test, sex_svm) %>%
ggplot(aes(x = sex, y = .pred_class, color = sex)) +
geom_jitter(size = 1, width = 0.2, height = 0.2) +
scale_color_manual(values = c("blue", "red")) +
theme_bw()2.2 펭귄종 분류모형
펭귄종(아델리, 젠투, 턱끈)을 분류하는 기계학습모형을 개발하기 전에 훈련/시험 데이터셋으로 나눈다.
2.2.1 다항회귀모형
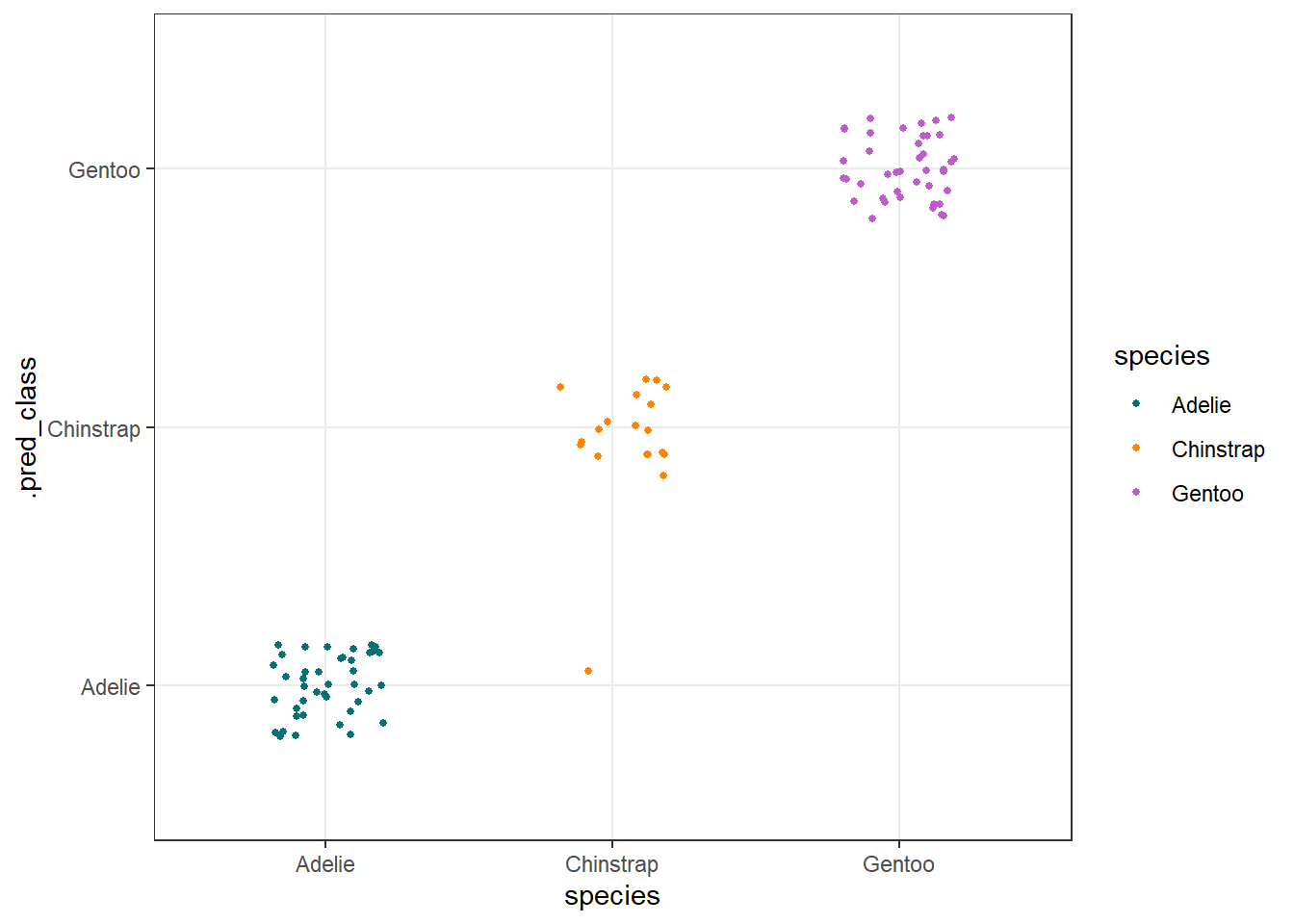
펭귄종(아델리, 젠투, 턱끈)을 분류하는 로지스틱 회귀모형으로 분류모형을 개발해보자.
코드
multinom_spec <- multinom_reg() %>%
set_mode("classification")
multinom_wflow <-
workflow(
species ~ sex + island + bill_length_mm + bill_depth_mm +
flipper_length_mm + body_mass_g,
multinom_spec
)
multinom_fit <- multinom_wflow %>% fit(data = penguins_train)
sex_predict <- predict(multinom_fit, penguins_test, type = "class")
bind_cols(penguins_test, sex_predict) %>%
ggplot(aes(x = species, y = .pred_class, color = species)) +
geom_jitter(size = 1, width = 0.2, height = 0.2) +
scale_color_manual(values = penguins_colors) +
theme_bw()2.2.2 Random Forest
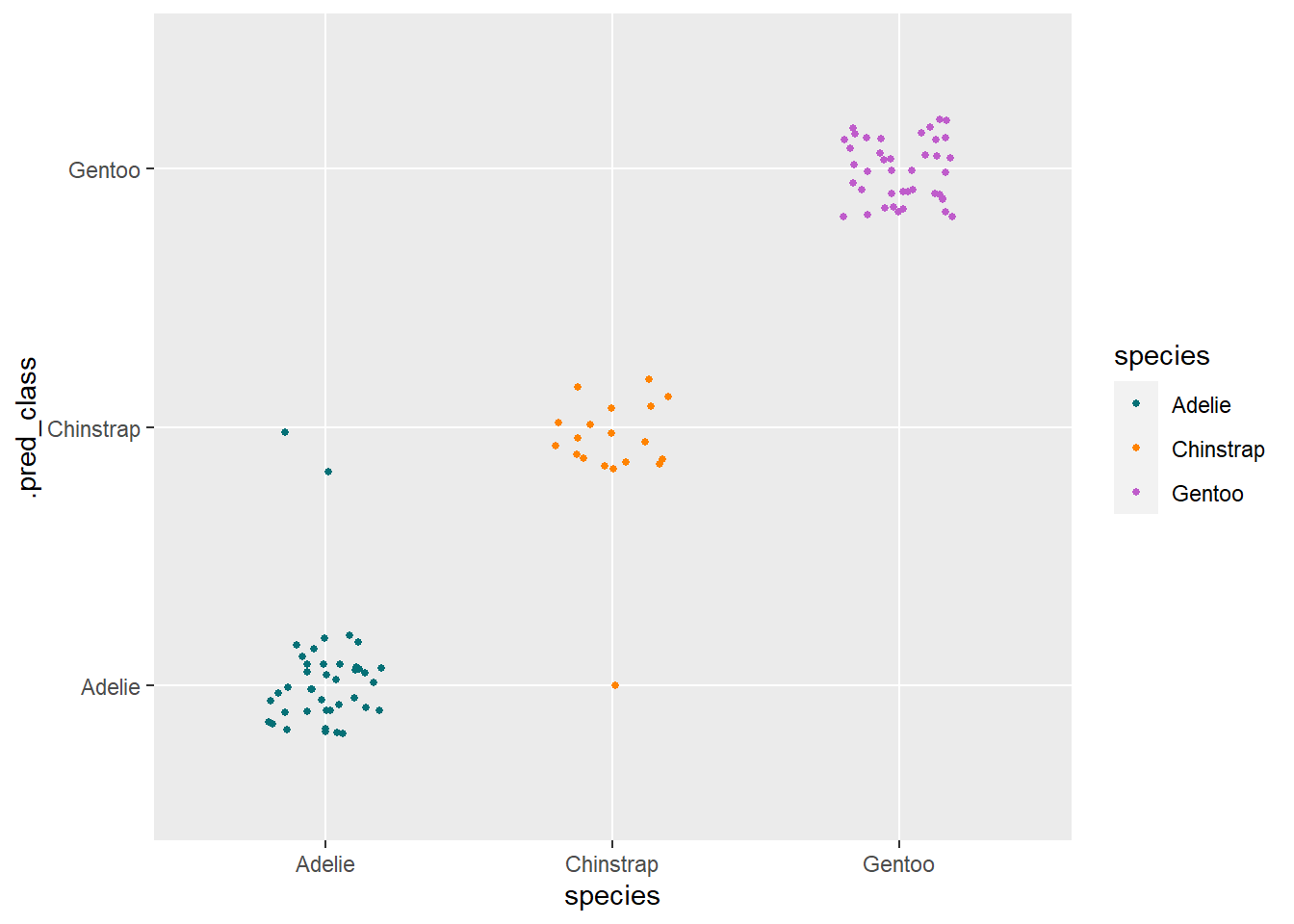
펭귄종(아델리, 젠투, 턱끈)을 분류하는 Random Forest 모형으로 분류모형을 개발해보자.
코드
rf_spec <- rand_forest() %>%
set_engine("ranger") %>%
set_mode("classification")
rf_wflow <-
workflow(
species ~ sex + island + bill_length_mm + bill_depth_mm +
flipper_length_mm + body_mass_g,
rf_spec
)
rf_fit <- rf_wflow %>% fit(data = penguins_train)
sex_rf <- predict(rf_fit, penguins_test, type = "class")
bind_cols(penguins_test, sex_rf) %>%
ggplot(aes(x = species, y = .pred_class, color = species)) +
geom_jitter(size = 1, width = 0.2, height = 0.2) +
scale_color_manual(values = penguins_colors) +
theme()3 고급 시각화
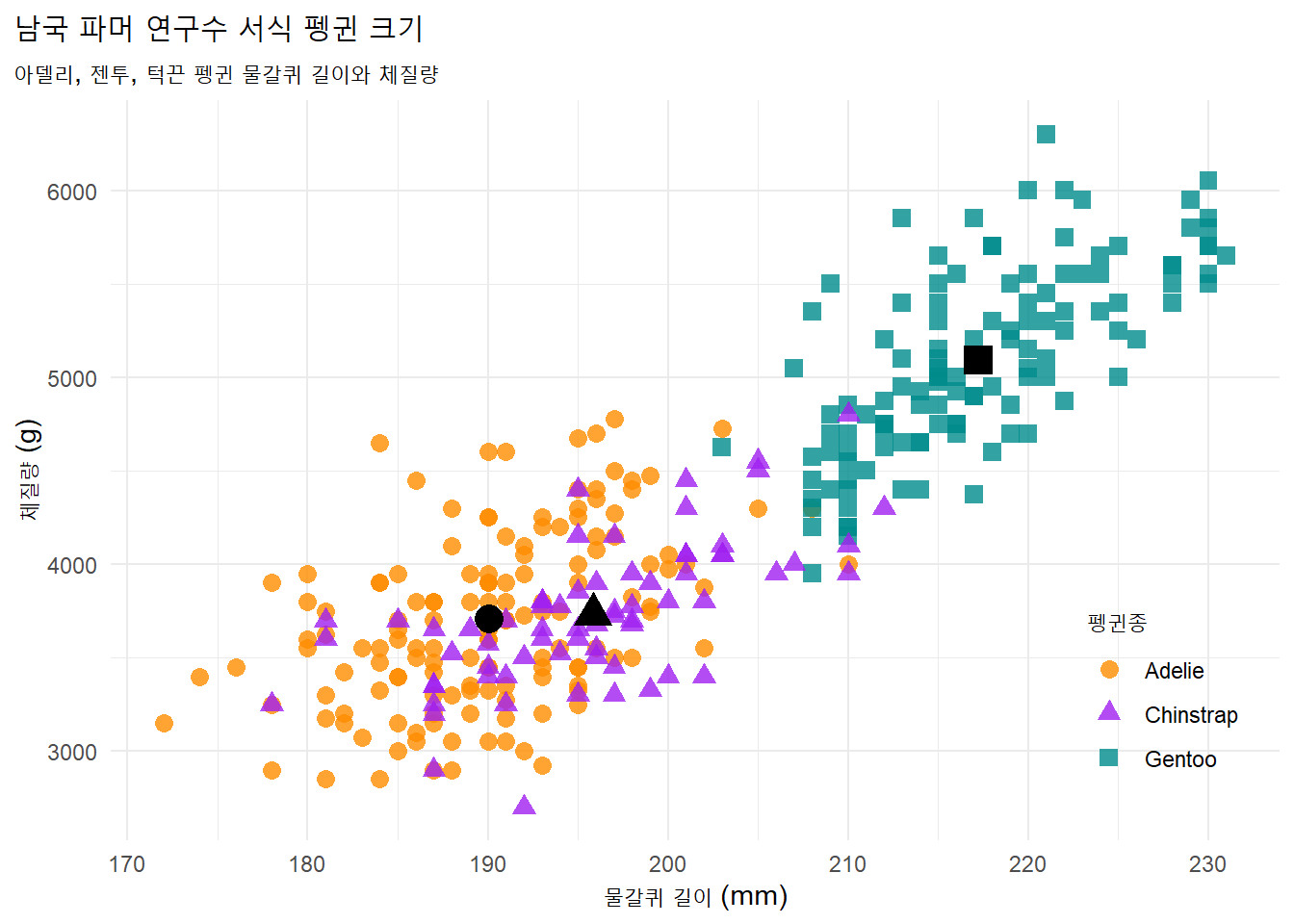
3.1 펭귄크기와 물칼퀴
코드
extrafont::loadfonts()
ggplot2::theme_set(ggplot2::theme_minimal(base_family = "MaruBuri"))
species_mean <- penguins %>%
group_by(species) %>%
summarise(flipper_mean = mean(flipper_length_mm),
body_mass_mean = mean(body_mass_g))
penguins %>%
ggplot(aes(x = flipper_length_mm, y = body_mass_g)) +
geom_point(aes(color = species, shape = species),
size = 3, alpha = 0.8) +
# 평균 추가
geom_point(data = species_mean, aes(x = flipper_mean, y = body_mass_mean, shape = species),
size = 5, color = "black", show.legend = FALSE) +
scale_color_manual(values = c("darkorange", "purple", "cyan4")) +
labs(title = "남국 파머 연구수 서식 펭귄 크기",
subtitle = "아델리, 젠투, 턱끈 펭귄 물갈퀴 길이와 체질량",
x = "물갈퀴 길이 (mm)",
y = "체질량 (g)",
color = "펭귄종",
shape = "펭귄종") +
theme(legend.position = c(0.9, 0.2),
plot.title.position = "plot",
plot.caption = element_text(hjust = 0, face= "italic"),
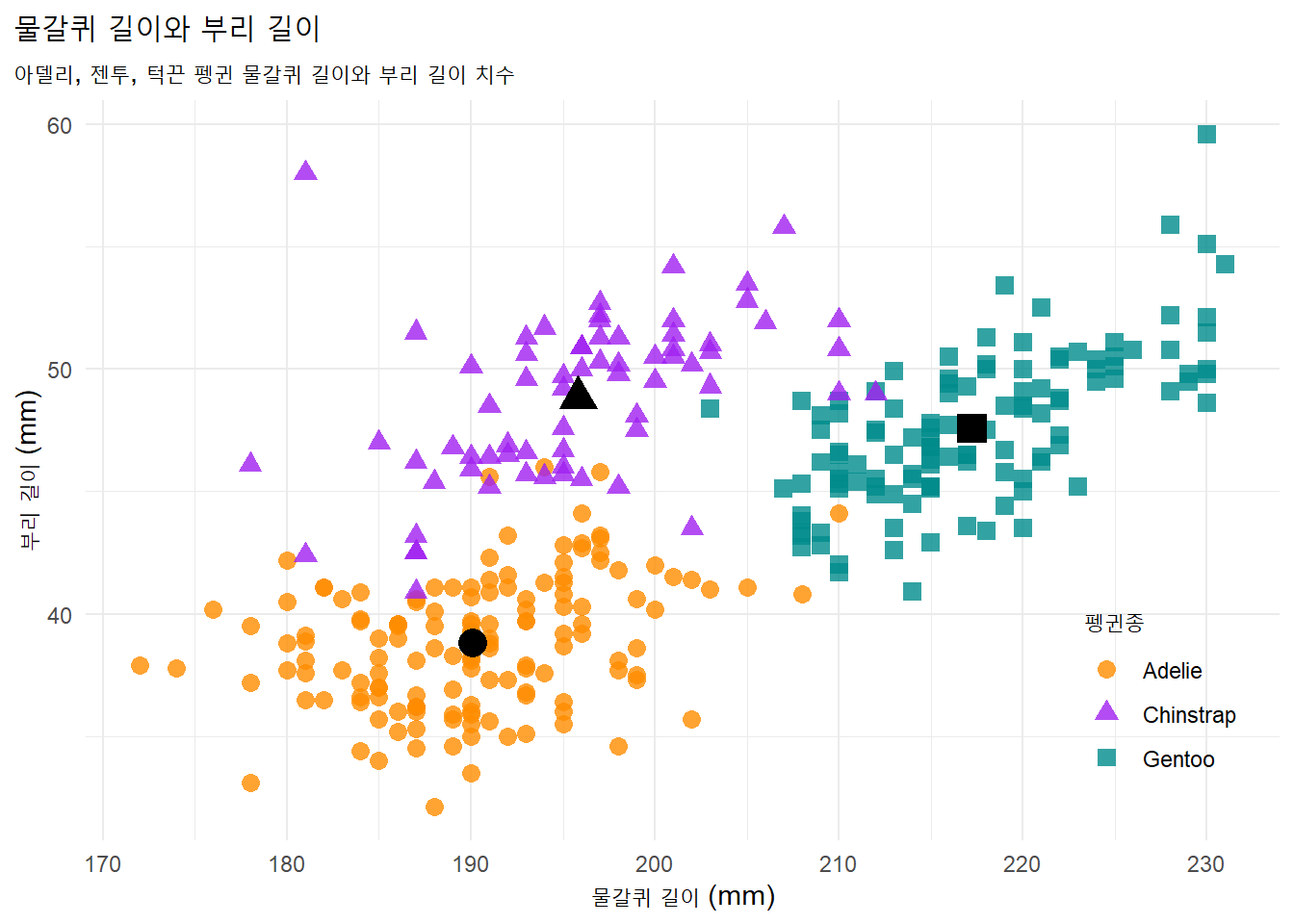
plot.caption.position = "plot") 3.2 물갈퀴와 부리 길이
코드
species_mean <- penguins %>%
group_by(species) %>%
summarise(flipper_mean = mean(flipper_length_mm),
bill_mean = mean(bill_length_mm))
penguins %>%
ggplot(aes(x = flipper_length_mm, y = bill_length_mm)) +
geom_point(aes(color = species, shape = species),
size = 3, alpha = 0.8) +
# 평균 추가
geom_point(data = species_mean, aes(x = flipper_mean, y = bill_mean, shape = species),
size = 5, color = "black", show.legend = FALSE) +
scale_color_manual(values = c("darkorange", "purple", "cyan4")) +
labs(title = "물갈퀴 길이와 부리 길이",
subtitle = "아델리, 젠투, 턱끈 펭귄 물갈퀴 길이와 부리 길이 치수",
x = "물갈퀴 길이 (mm)",
y = "부리 길이 (mm)",
color = "펭귄종",
shape = "펭귄종") +
theme(legend.position = c(0.9, 0.2),
plot.title.position = "plot",
plot.caption = element_text(hjust = 0, face= "italic"),
plot.caption.position = "plot") 3.3 분포와 Facet
코드
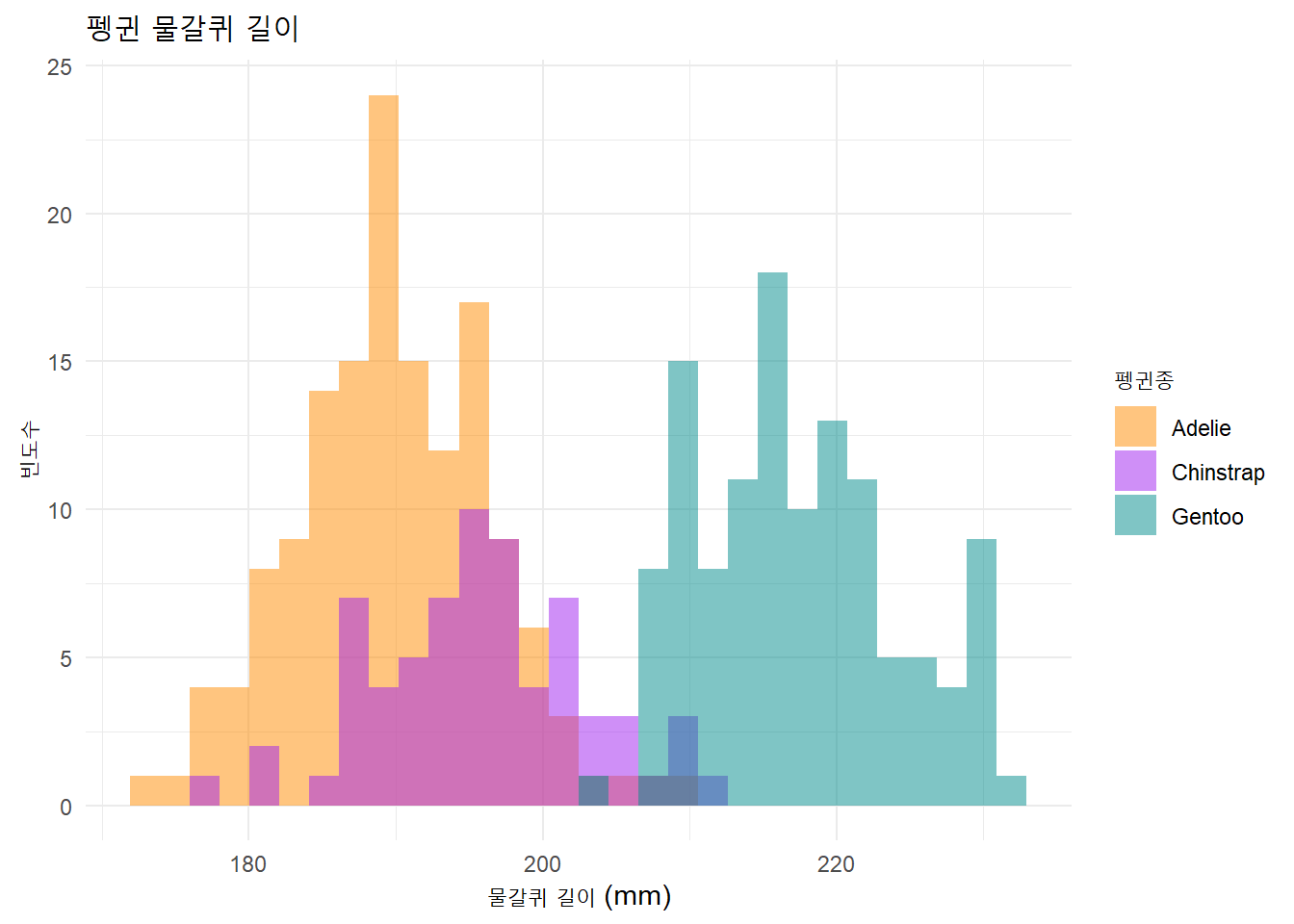
ggplot(data = penguins, aes(x = flipper_length_mm)) +
geom_histogram(aes(fill = species),
alpha = 0.5,
position = "identity") +
scale_fill_manual(values = c("darkorange","purple","cyan4")) +
labs(x = "물갈퀴 길이 (mm)",
y = "빈도수",
title = "펭귄 물갈퀴 길이",
fill = "펭귄종")코드
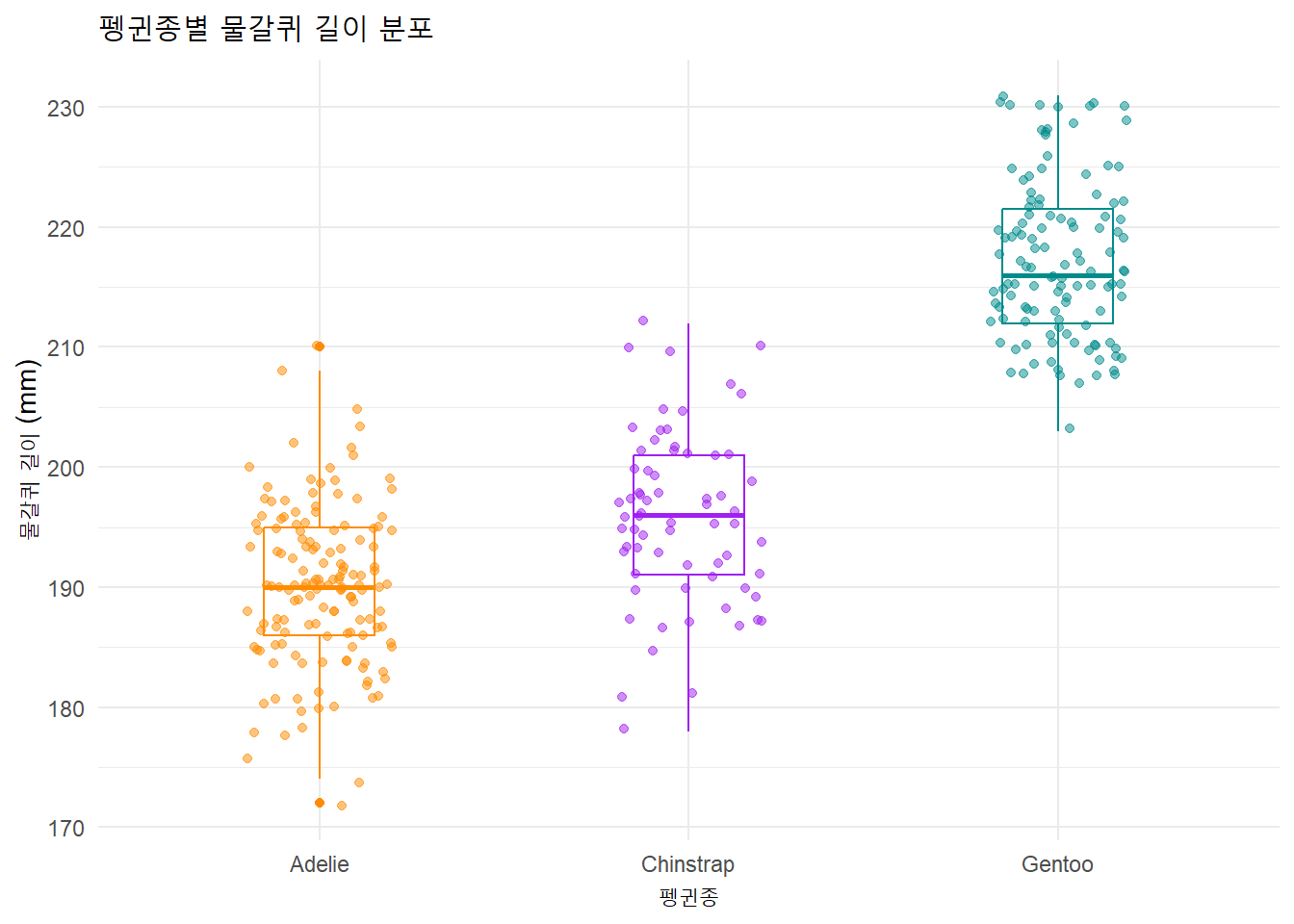
ggplot(data = penguins, aes(x = species, y = flipper_length_mm)) +
geom_boxplot(aes(color = species), width = 0.3, show.legend = FALSE) +
geom_jitter(aes(color = species), alpha = 0.5, show.legend = FALSE,
position = position_jitter(width = 0.2, seed = 0)) +
scale_color_manual(values = c("darkorange","purple","cyan4")) +
labs(x = "펭귄종",
y = "물갈퀴 길이 (mm)",
title = "펭귄종별 물갈퀴 길이 분포")코드
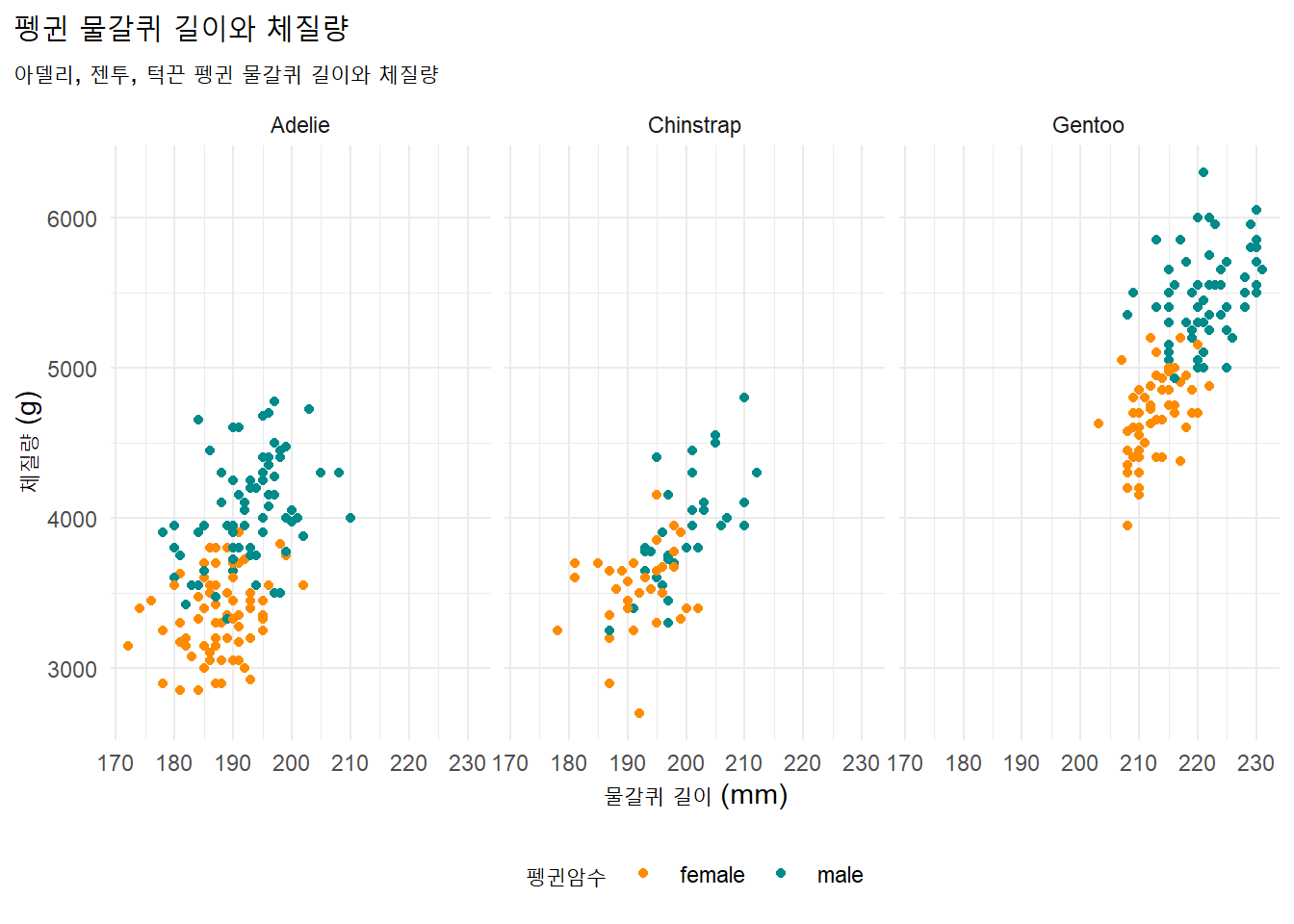
ggplot(penguins, aes(x = flipper_length_mm, y = body_mass_g)) +
geom_point(aes(color = sex)) +
scale_color_manual(values = c("darkorange","cyan4"), na.translate = FALSE) +
labs(title = "펭귄 물갈퀴 길이와 체질량",
subtitle = "아델리, 젠투, 턱끈 펭귄 물갈퀴 길이와 체질량",
x = "물갈퀴 길이 (mm)",
y = "체질량 (g)",
color = "펭귄암수") +
theme(legend.position = "bottom",
plot.title.position = "plot",
plot.caption = element_text(hjust = 0, face= "italic"),
plot.caption.position = "plot") +
facet_wrap(~species)4 심슨 역설
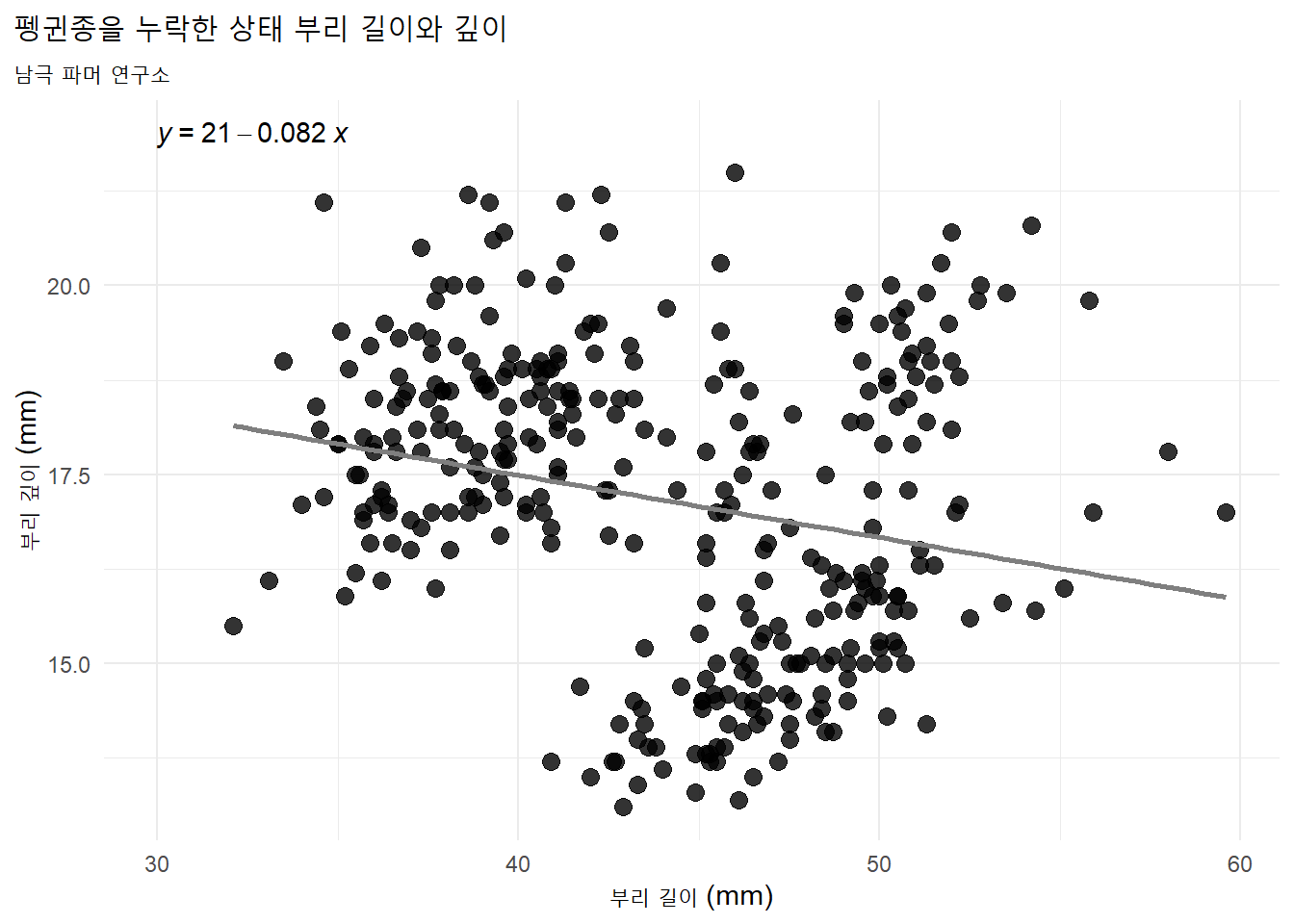
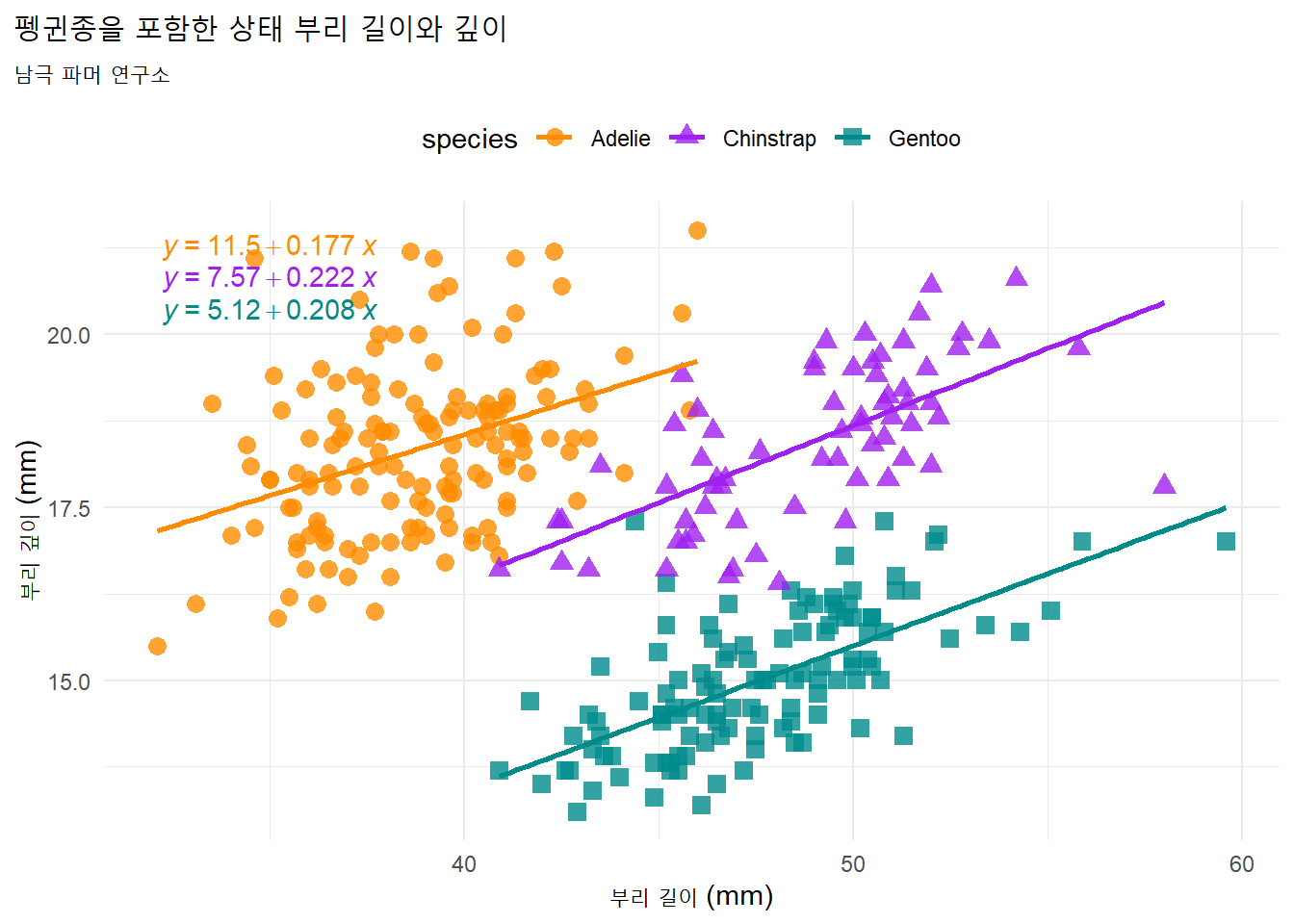
심슨의 역설(Simpson’s Paradox)은 데이터를 취합할 때 의미 있는 변수를 생략하면 변수 간에 관찰되는 추세가 역전되는 데이터 현상이다.
부리의 길이와 깊이는 전체적으로 음의 상관관계를 보이지만, 종을 포함하면 이러한 추세가 반전되어 종 내에서는 부리 길이와 부리 깊이 사이에 양의 상관관계가 뚜렷하게 나타난다.
펭귄종 누락 시각화
코드
library(ggpubr)
penguins %>%
ggplot(aes(x = bill_length_mm, y = bill_depth_mm)) +
geom_point(size = 3, alpha = 0.8) +
labs(title = "펭귄종을 누락한 상태 부리 길이와 깊이",
subtitle = "남극 파머 연구소",
x = "부리 길이 (mm)",
y = "부리 깊이 (mm)") +
theme(plot.title.position = "plot",
plot.caption = element_text(hjust = 0, face= "italic"),
plot.caption.position = "plot") +
geom_smooth(method = "lm", se = FALSE, color = "gray50") +
stat_regline_equation(label.x = 30, label.y = 22)펭귄종 포함 시각화
코드
library(ggpmisc)
penguins %>%
ggplot(aes(x = bill_length_mm, y = bill_depth_mm,
color = species, shape = species)) +
geom_point(size = 3, alpha = 0.8) +
scale_color_manual(values = c("darkorange","purple","cyan4")) +
stat_poly_line(formula = y ~ x, se = FALSE) +
stat_poly_eq(aes(label = after_stat(eq.label)), formula = y ~ x) +
labs(title = "펭귄종을 포함한 상태 부리 길이와 깊이",
subtitle = "남극 파머 연구소",
x = "부리 길이 (mm)",
y = "부리 깊이 (mm)") +
theme(plot.title.position = "plot",
plot.caption = element_text(hjust = 0, face= "italic"),
plot.caption.position = "plot",
legend.position = "top") 5 다변량 분석
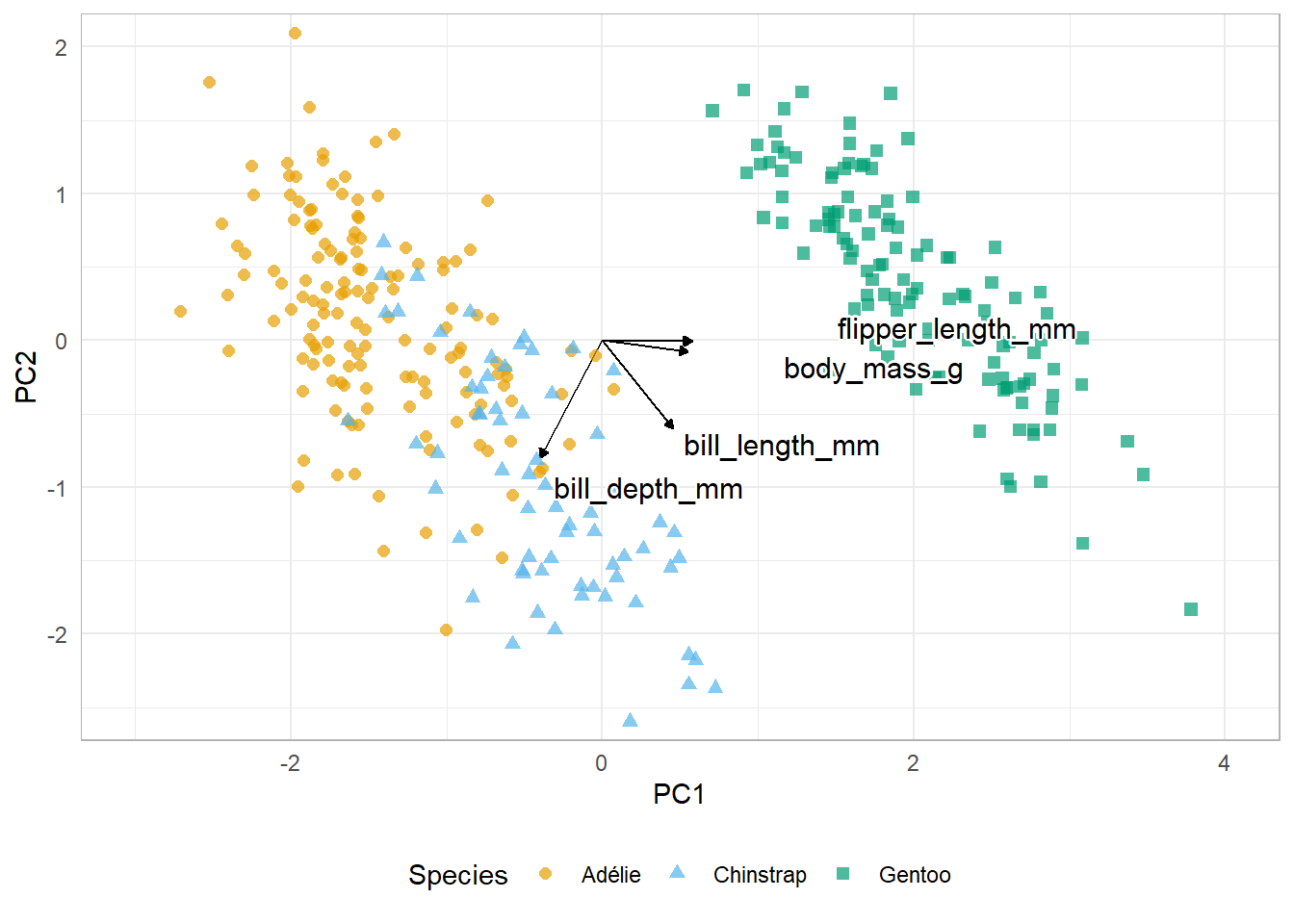
5.1 주성분 분석
주성분 분석(PCA)은 다변량 데이터의 패턴을 탐색하는 데 일반적으로 사용되는 차원 축소 방법이다. (Allison M. Horst, 2022)
코드
# Omit year
penguins_noyr <- penguins %>%
select(-year) %>%
mutate(species = as.character(species)) %>%
mutate(species = case_when(
species == "Adelie" ~ "Adélie",
TRUE ~ species
)) %>%
mutate(species = as.factor(species))
penguin_recipe <-
recipe(~., data = penguins_noyr) %>%
update_role(species, island, sex, new_role = "id") %>%
step_naomit(all_predictors()) %>%
step_normalize(all_predictors()) %>%
step_pca(all_predictors(), id = "pca") %>%
prep()
penguin_pca <-
penguin_recipe %>%
tidy(id = "pca")
penguin_percvar <- penguin_recipe %>%
tidy(id = "pca", type = "variance") %>%
dplyr::filter(terms == "percent variance")
# Make the penguins PCA biplot:
# Get pca loadings into wider format
pca_wider <- penguin_pca %>%
tidyr::pivot_wider(names_from = component, id_cols = terms)
# define arrow style:
arrow_style <- arrow(length = unit(.05, "inches"),
type = "closed")
penguins_juiced <- juice(penguin_recipe)
# Make the penguins PCA biplot:
pca_plot <-
penguins_juiced %>%
ggplot(aes(PC1, PC2)) +
coord_cartesian(
xlim = c(-3, 4),
ylim = c(-2.5, 2)) +
paletteer::scale_color_paletteer_d("colorblindr::OkabeIto") +
guides(color = guide_legend("Species"),
shape = guide_legend("Species")) +
theme(legend.position = "bottom",
panel.border = element_rect(color = "gray70", fill = NA))
# For positioning (above):
# 1: bill_length
# 2: bill_depth
# 3: flipper length
# 4: body mass
penguins_biplot <- pca_plot +
geom_segment(data = pca_wider,
aes(xend = PC1, yend = PC2),
x = 0,
y = 0,
arrow = arrow_style) +
geom_point(aes(color = species, shape = species),
alpha = 0.7,
size = 2) +
shadowtext::geom_shadowtext(data = pca_wider,
aes(x = PC1, y = PC2, label = terms),
nudge_x = c(0.7,0.7,1.7,1.2),
nudge_y = c(-0.1,-0.2,0.1,-0.1),
size = 4,
color = "black",
bg.color = "white")
penguins_biplot
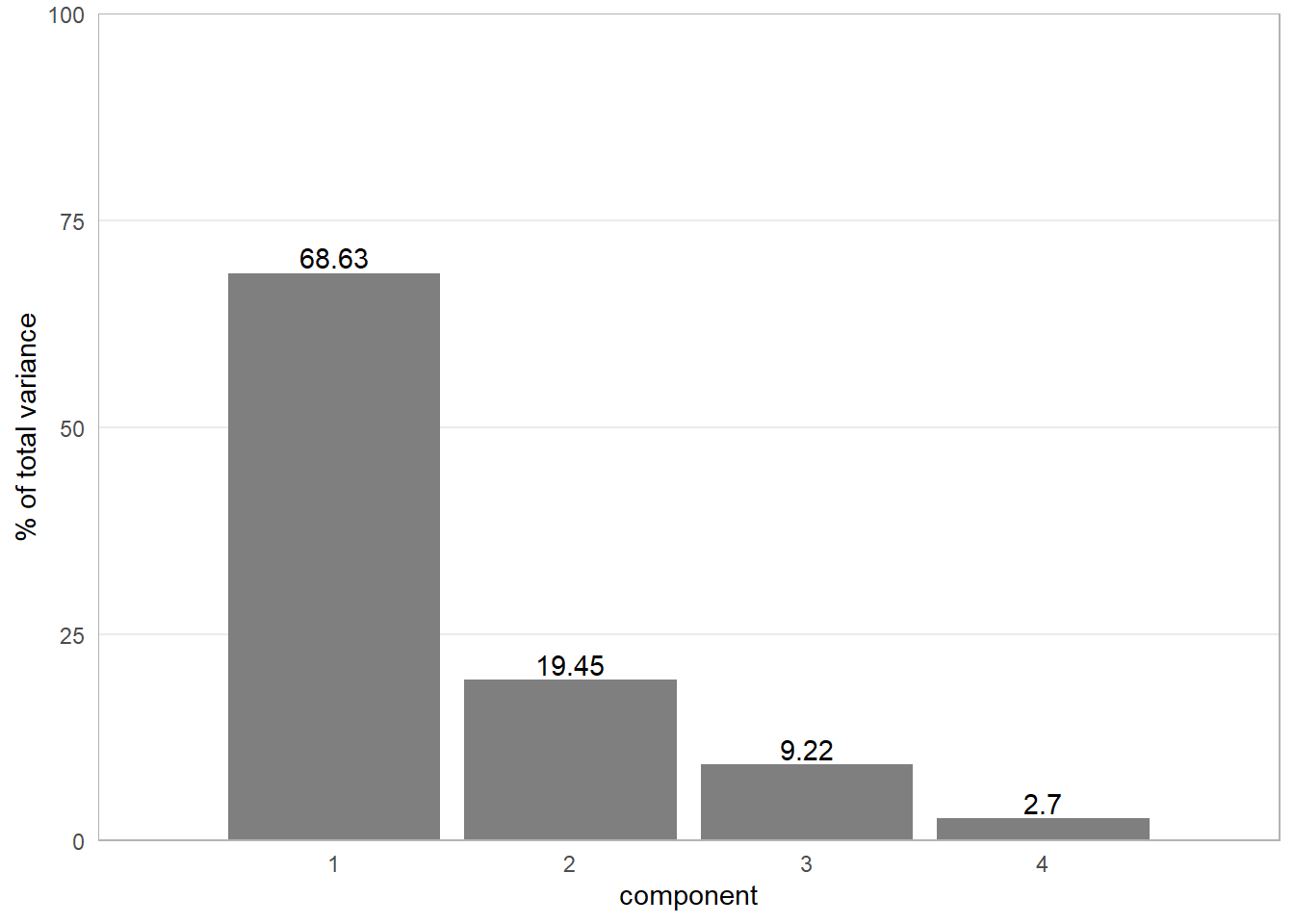
penguin_screeplot_base <- penguin_percvar %>%
ggplot(aes(x = component, y = value)) +
scale_x_continuous(limits = c(0, 5), breaks = c(1,2,3,4), expand = c(0,0)) +
scale_y_continuous(limits = c(0,100), expand = c(0,0)) +
ylab("% of total variance") +
theme(panel.border = element_rect(color = "gray70", fill = NA),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank(),
panel.grid.minor.y = element_blank())
penguin_screeplot <- penguin_screeplot_base +
geom_col(fill = "gray50") +
geom_text(aes(label = round(value,2)), vjust=-0.25)
penguin_screeplot5.2 군집분석
k-평균(K-Means) 비지도학습 군집분석은 기계학습 및 분류에 대한 일반적이고 인기 있는 알고리즘이다.
코드
## ---- kmeans ---------------------------------------------------------
# TWO VARIABLE k-means comparison
# Penguins: Bill length vs. bill depth
pb_species <- penguins %>%
select(species, starts_with("bill")) %>%
drop_na() %>%
mutate(species = as.character(species)) %>%
mutate(species = case_when(
species == "Adelie" ~ "Adélie",
TRUE ~ species
)) %>%
mutate(species = as.factor(species))
# Prep penguins for k-means:
pb_nospecies <- pb_species %>%
select(-species) %>%
recipe() %>%
step_normalize(all_numeric()) %>%
prep() %>%
juice()
# Perform k-means on penguin bill dimensions (k = 3, w/20 centroid starts)
# Save augmented data
set.seed(100)
pb_clust <-
pb_nospecies %>%
kmeans(centers = 3, nstart = 20) %>%
broom::augment(pb_species)
# Get counts in each cluster by species
pb_clust_n <- pb_clust %>%
count(species, .cluster) %>%
pivot_wider(names_from = species, values_from = n, names_sort = TRUE) %>%
arrange(.cluster) %>%
replace_na(list(`Adelie` = 0))
# Plot penguin k-means clusters:
# make a base plot b/c https://github.com/plotly/plotly.R/issues/1942
pb_kmeans_base <-
pb_clust %>%
ggplot(aes(x = bill_length_mm, y = bill_depth_mm)) +
paletteer::scale_color_paletteer_d("colorblindr::OkabeIto") +
paletteer::scale_fill_paletteer_d("colorblindr::OkabeIto") +
scale_x_continuous(limits = c(30, 60),
breaks = c(30, 40, 50, 60)) +
theme(legend.position = "bottom",
panel.border = element_rect(fill = NA, color = "gray70")) +
labs(x = "Bill length (mm)",
y = "Bill depth (mm)",
color = "Species")
# ggpubr::stat_chull(aes(fill = .cluster, color = .cluster),
# alpha = 0.5, geom = "polygon", show.legend = FALSE)
pb_kmeans_gg <- pb_kmeans_base +
geom_text(aes(label = .cluster,
color = species),
key_glyph = draw_key_rect,
check_overlap = TRUE)
pb_kmeans_plotly <- pb_kmeans_base +
geom_text(aes(label = .cluster,
color = species,
text = paste("Species: ", species,
"\nCluster: ", .cluster,
"\nBill length (mm): ", bill_length_mm,
"\nBill depth (mm): ", bill_depth_mm)
),
size = 3)
plotly::ggplotly(pb_kmeans_plotly, height = 300, tooltip = "text")