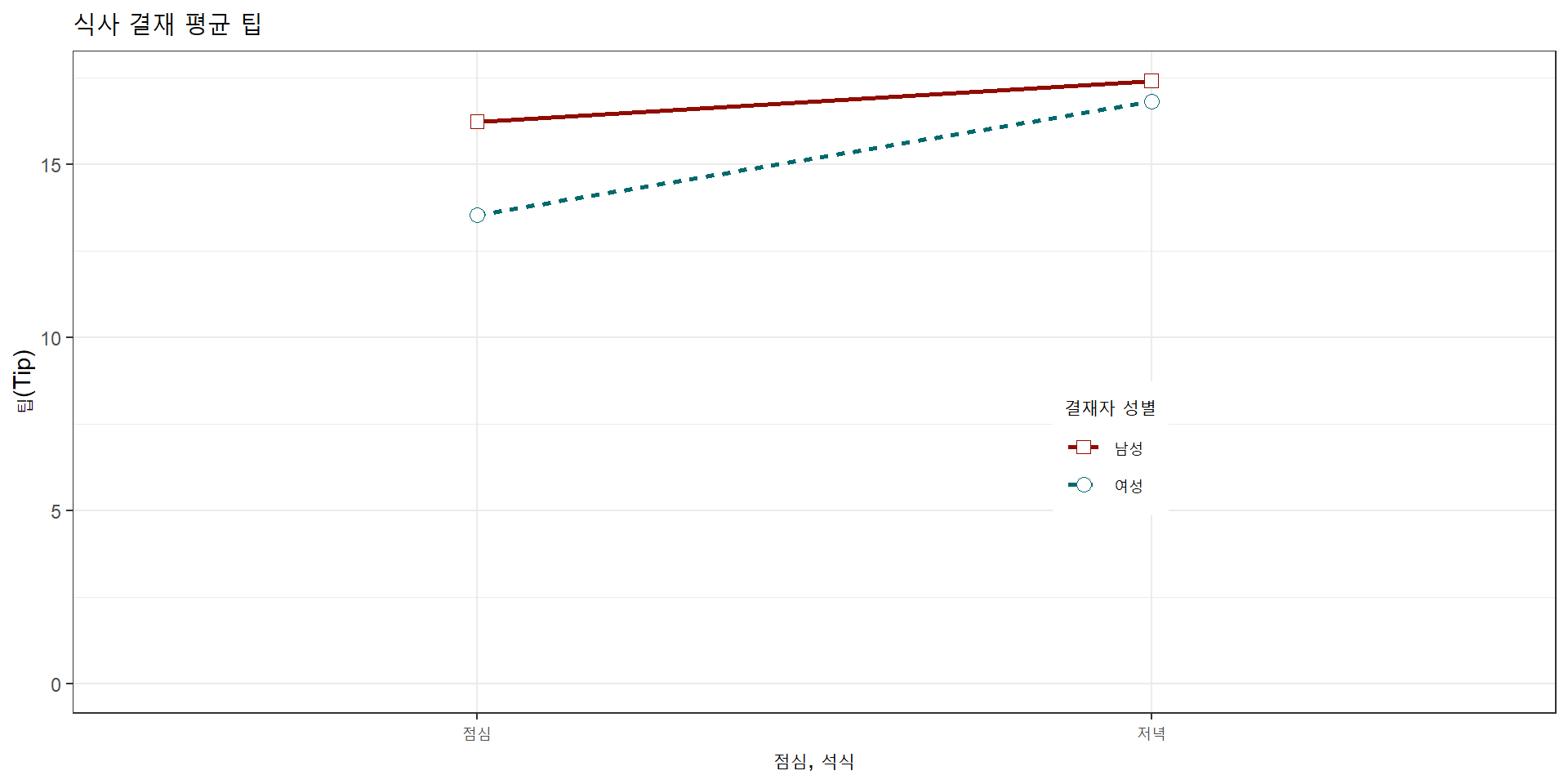데이터 사이언스와 디자인
디자인과 아키텍쳐의 중요성
Invalid Date
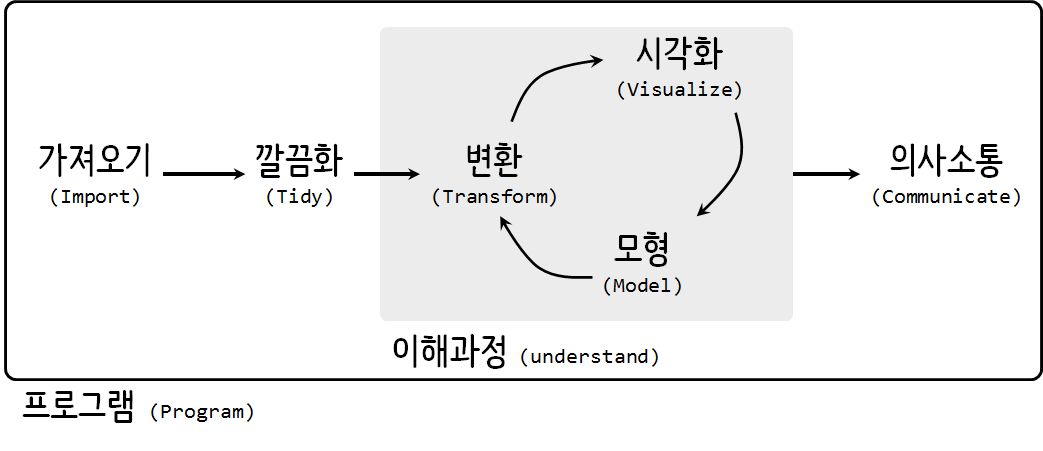
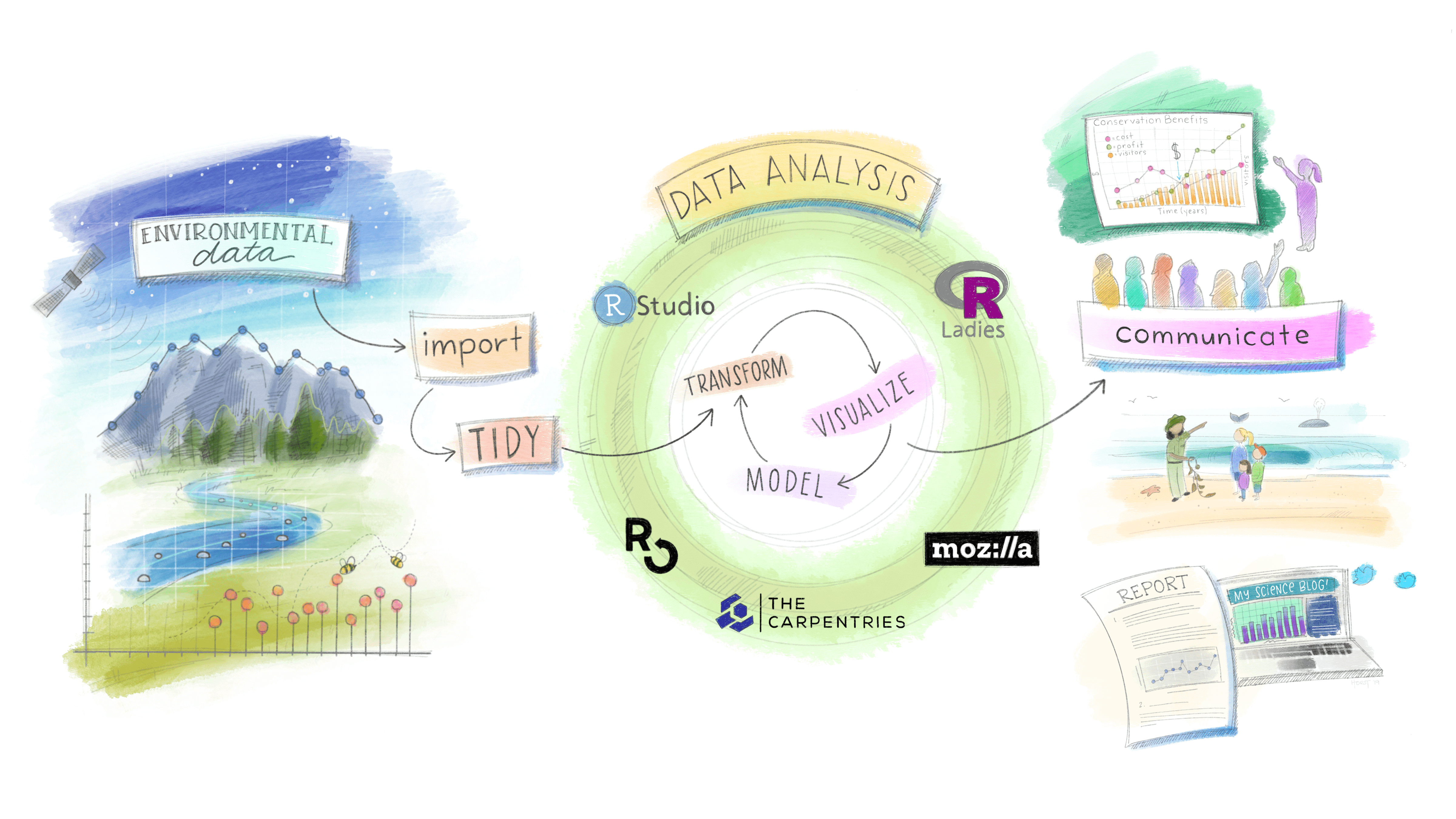
데이터 과학
데이터 사이언스
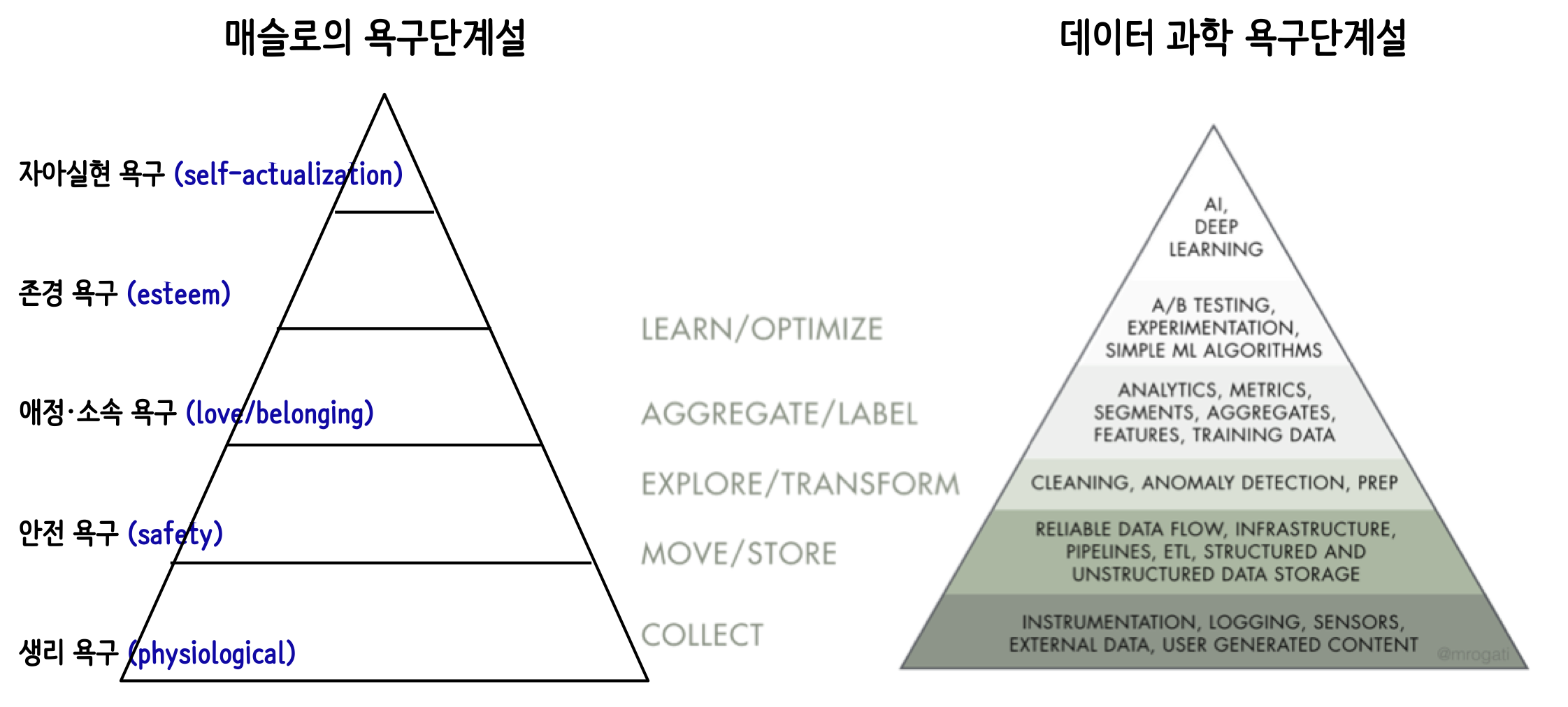
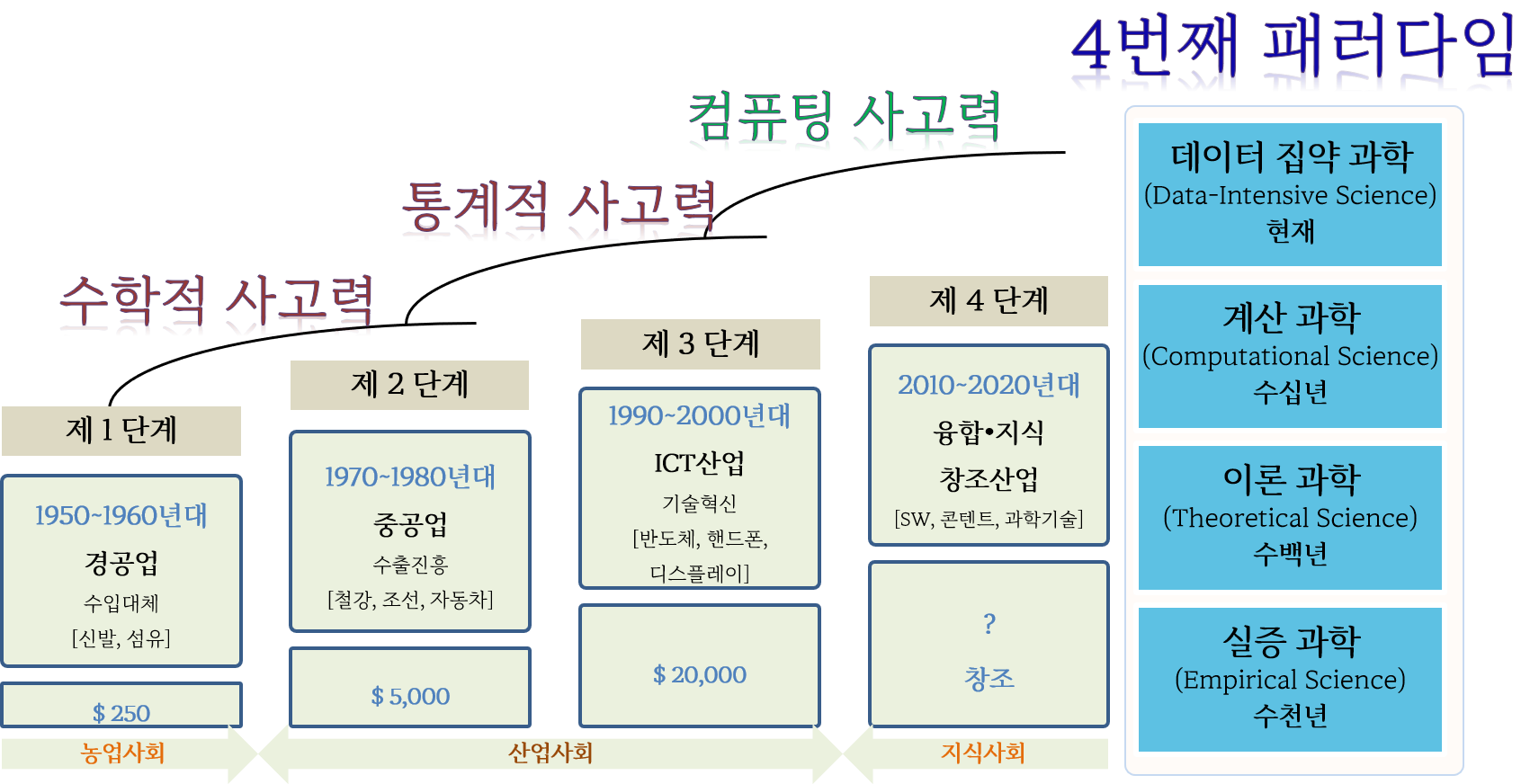
데이터 과학 욕구단계설
문제
데이터 사이언스 - 출판
복잡성
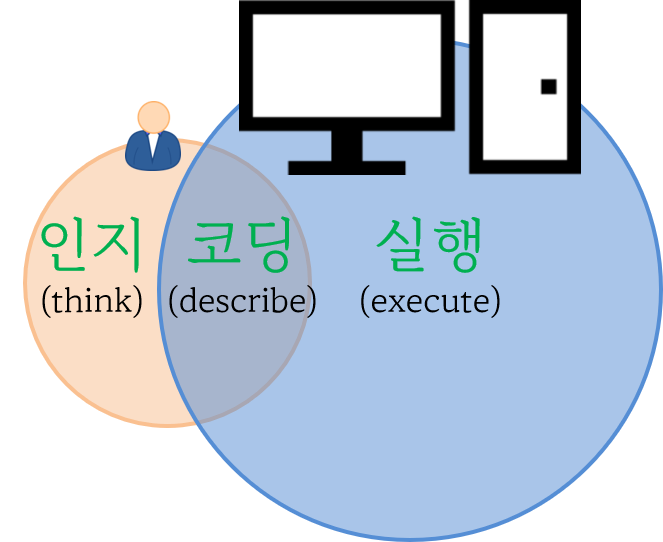
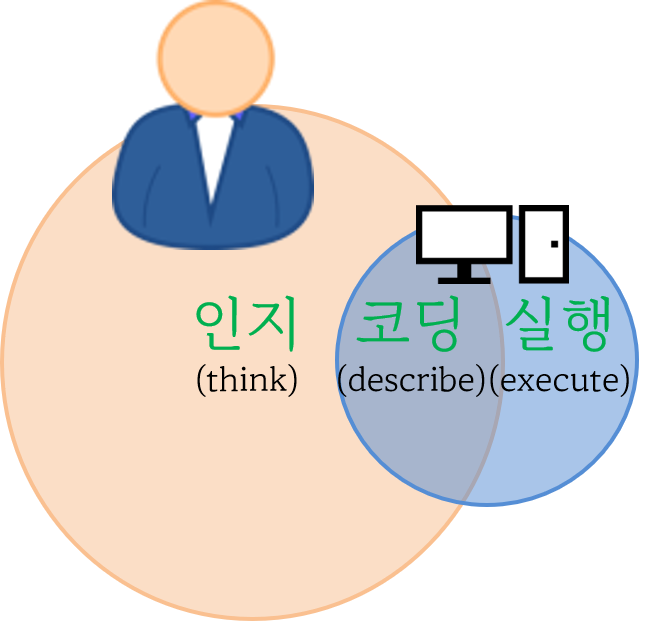
사람 대 기계
해법
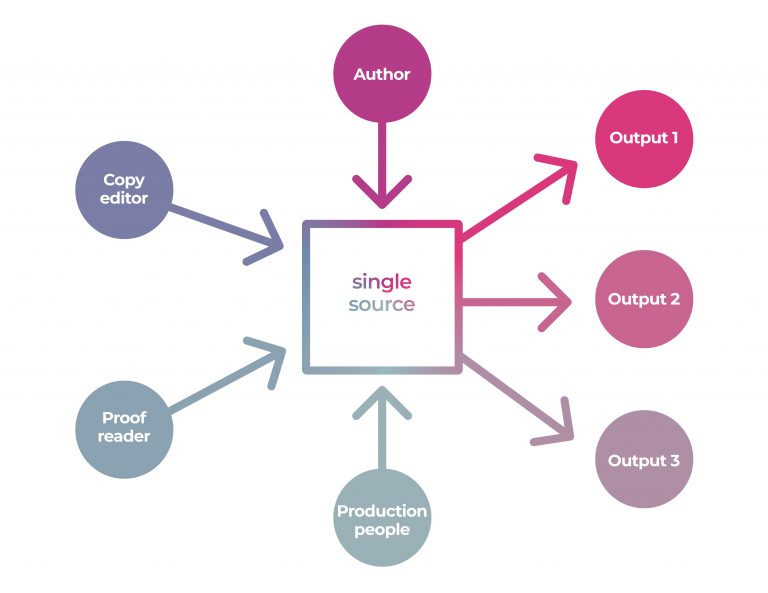
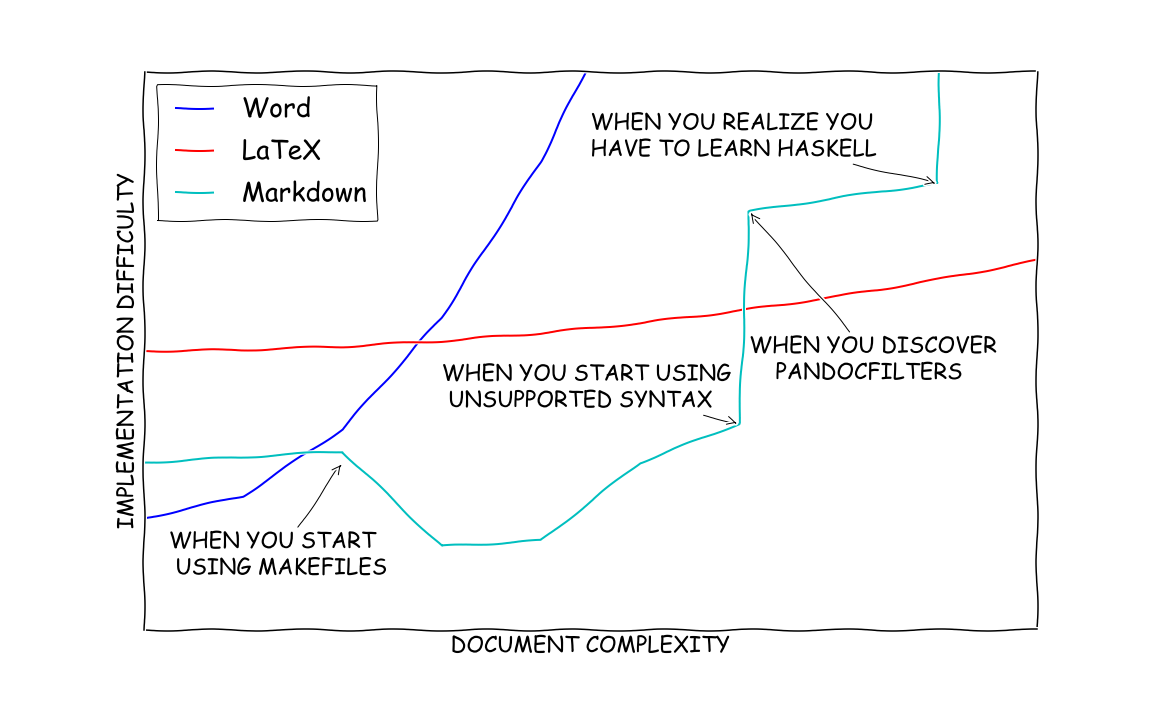
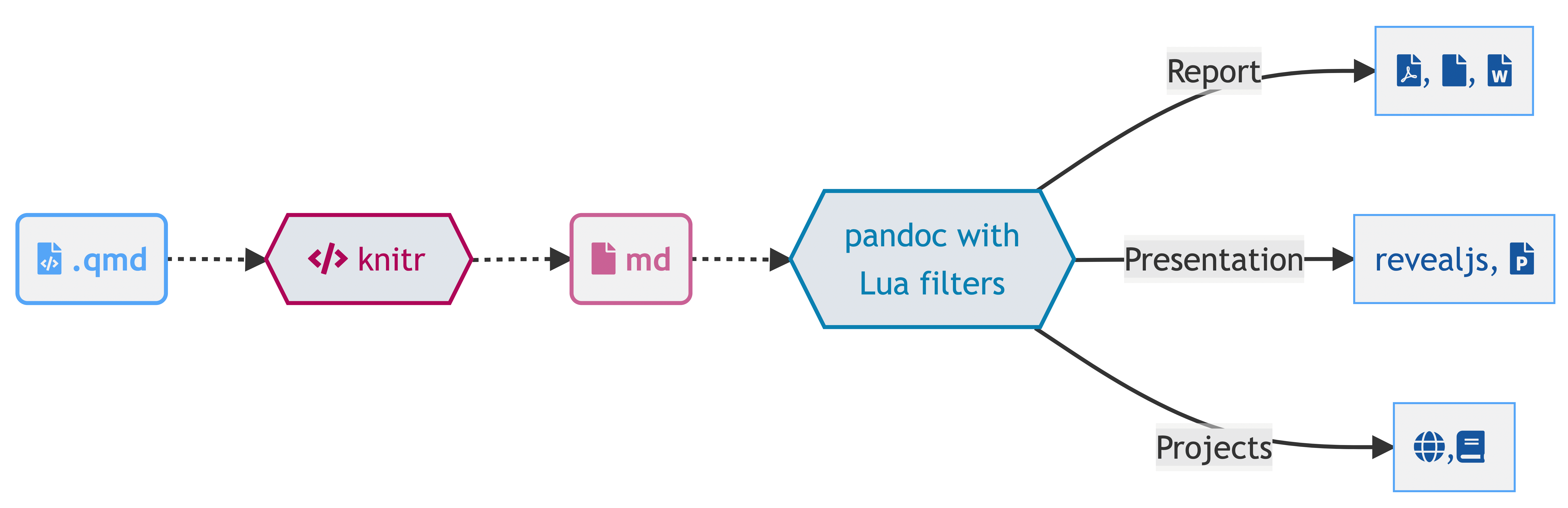
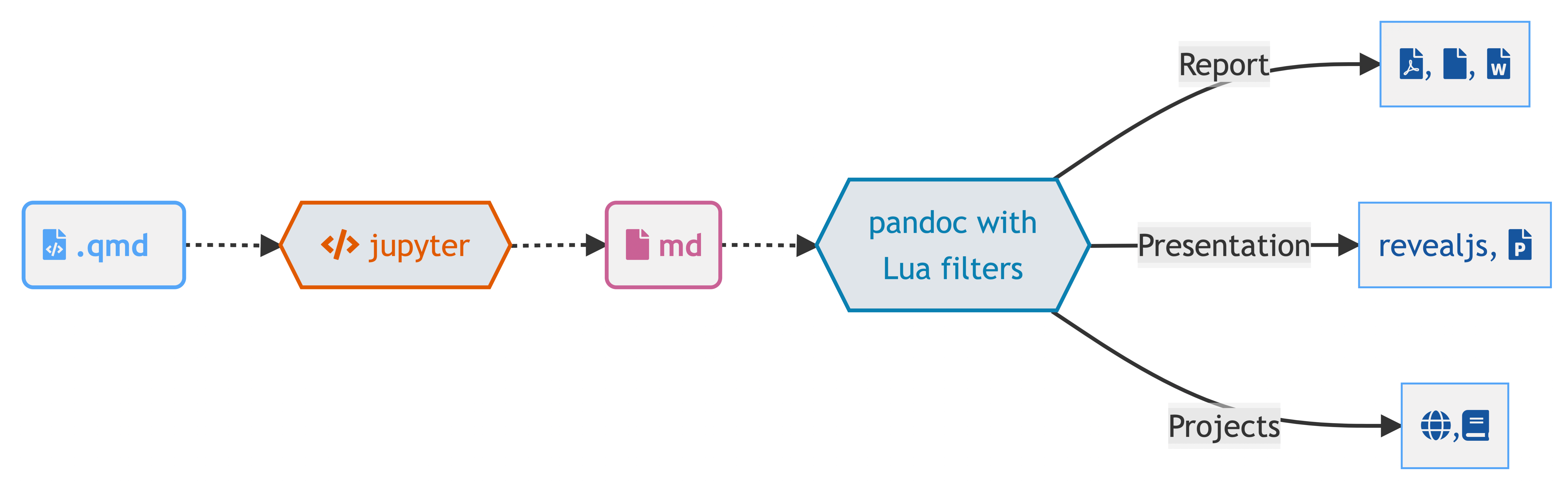
Quarto
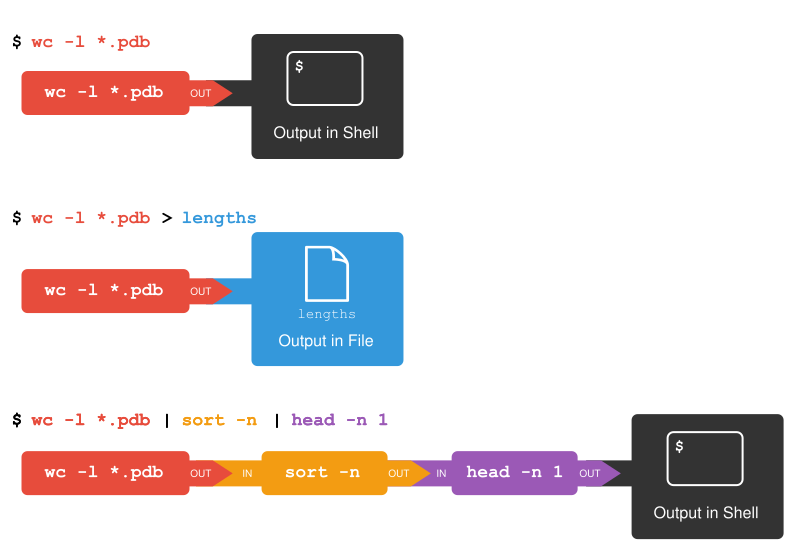
파이프 & 필터
시각화 - 그래프 문법 1
R 코드
bill_df %>%
ggplot(aes(x=time, y=total_bill, group=sex, shape=sex, colour=sex)) +
geom_line(aes(linetype=sex), size=1) +
geom_point(size=3, fill="white") +
expand_limits(y=0) +
scale_colour_hue(name="결재자 성별", l=30) +
scale_shape_manual(name="결재자 성별", values=c(22,21)) +
scale_linetype_discrete(name="결재자 성별") +
xlab("점심, 석식") + ylab("팁(Tip)") +
ggtitle("식사 결재 평균 팁") +
theme_bw() +
theme(legend.position=c(.7, .4))R 코드
bill_mat <- matrix( bill_df$total_bill,
nrow = 2,
byrow=TRUE,
dimnames = list(c("여성", "남성"), c("점심", "저녁"))
)
mf_col <- c("#3CC3BD", "#FD8210")
barplot(bill_mat, beside = TRUE, border=NA, col=mf_col)
legend("topleft", row.names(bill_mat), pch=15, col=mf_col)
par(cex=1.2, cex.axis=1.1)
matplot(bill_mat, type="b", lty=1, pch=19, col=mf_col,
cex=1.5, lwd=3, las=1, bty="n", xaxt="n",
xlim=c(0.7, 2.2), ylim=c(12,18), ylab="",
main="식사 결재 평균 팁", yaxt="n")
axis(2, at=axTicks(2), labels=sprintf("$%s", axTicks(2)),
las=1, cex.axis=0.8, col=NA, line = -0.5)
grid(NA, NULL, lty=3, lwd=1, col="#000000")
abline(v=c(1,2), lty=3, lwd=1, col="#000000")
mtext("점심", side=1, at=1)
mtext("저녁", side=1, at=2)
text(1.5, 17.3, "남성", srt=8, font=3)
text(1.5, 15.1, "여성", srt=33, font=3)디자인 - 코드
날짜와 시간별로 총 빈도수와 출발연착 평균 시간을 구한다. 단, 총빈도수가 10 회 미만인 것은 제외하고 출발연착 시간의 결측값을 제거하고 계산한다.
-
flights데이터프레임에서 - 출발 연착시간(
dep_delay) 칼럼에서 결측값이 없는 (!is.na()) 행을 필터링 하고 - 날짜별(
date), 시간별(hour) 그룹을 묶어 - 평균 출발 연착시간을 계산하고 총빈도수 총계를 내고
- 총빈도수가 10회 이상인 날짜와 시간을 찾아내시오
디자인 - 데이터
- WHO 결핵 원데이터
- WHO 에서 년도별, 국가별, 연령별, 성별, 진단방법별 결핵 발병사례 조사 통계 데이터
- 진단방법
-
relstands for cases of relapse -
epstands for cases of extrapulmonary TB -
snstands for cases of pulmonary TB that could not be diagnosed by a pulmonary smear (smear negative) -
spstands for cases of pulmonary TB that could be diagnosed by a pulmonary smear (smear positive)
-
- 연령
- 014 = 0 – 14 years old
- 1524 = 15 – 24 years old
- 2534 = 25 – 34 years old
- 3544 = 35 – 44 years old
- 4554 = 45 – 54 years old
- 5564 = 55 – 64 years old
- 65 = 65 or older
- 성별
- males (m)
- females (f)
library(tidyverse)
tidyr::who
#> # A tibble: 7,240 × 60
#> country iso2 iso3 year new_sp_m014 new_sp_m1524 new_sp_m2534 new_sp_m3544
#> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Afghani… AF AFG 1980 NA NA NA NA
#> 2 Afghani… AF AFG 1981 NA NA NA NA
#> 3 Afghani… AF AFG 1982 NA NA NA NA
#> 4 Afghani… AF AFG 1983 NA NA NA NA
#> 5 Afghani… AF AFG 1984 NA NA NA NA
#> 6 Afghani… AF AFG 1985 NA NA NA NA
#> 7 Afghani… AF AFG 1986 NA NA NA NA
#> 8 Afghani… AF AFG 1987 NA NA NA NA
#> 9 Afghani… AF AFG 1988 NA NA NA NA
#> 10 Afghani… AF AFG 1989 NA NA NA NA
#> # ℹ 7,230 more rows
#> # ℹ 52 more variables: new_sp_m4554 <dbl>, new_sp_m5564 <dbl>,
#> # new_sp_m65 <dbl>, new_sp_f014 <dbl>, new_sp_f1524 <dbl>,
#> # new_sp_f2534 <dbl>, new_sp_f3544 <dbl>, new_sp_f4554 <dbl>,
#> # new_sp_f5564 <dbl>, new_sp_f65 <dbl>, new_sn_m014 <dbl>,
#> # new_sn_m1524 <dbl>, new_sn_m2534 <dbl>, new_sn_m3544 <dbl>,
#> # new_sn_m4554 <dbl>, new_sn_m5564 <dbl>, new_sn_m65 <dbl>, …who %>%
pivot_longer(
cols = new_sp_m014:newrel_f65,
names_to = "key",
values_to = "cases",
values_drop_na = TRUE
) %>%
mutate(
key = stringr::str_replace(key, "newrel", "new_rel")
) %>%
separate(key, c("new", "var", "sexage")) %>%
select(-new, -iso2, -iso3) %>%
separate(sexage, c("sex", "age"), sep = 1)#> # A tibble: 76,046 × 6
#> country year var sex age cases
#> <chr> <dbl> <chr> <chr> <chr> <dbl>
#> 1 Afghanistan 1997 sp m 014 0
#> 2 Afghanistan 1997 sp m 1524 10
#> 3 Afghanistan 1997 sp m 2534 6
#> 4 Afghanistan 1997 sp m 3544 3
#> 5 Afghanistan 1997 sp m 4554 5
#> 6 Afghanistan 1997 sp m 5564 2
#> 7 Afghanistan 1997 sp m 65 0
#> 8 Afghanistan 1997 sp f 014 5
#> 9 Afghanistan 1997 sp f 1524 38
#> 10 Afghanistan 1997 sp f 2534 36
#> # ℹ 76,036 more rows디자인과 아키텍처
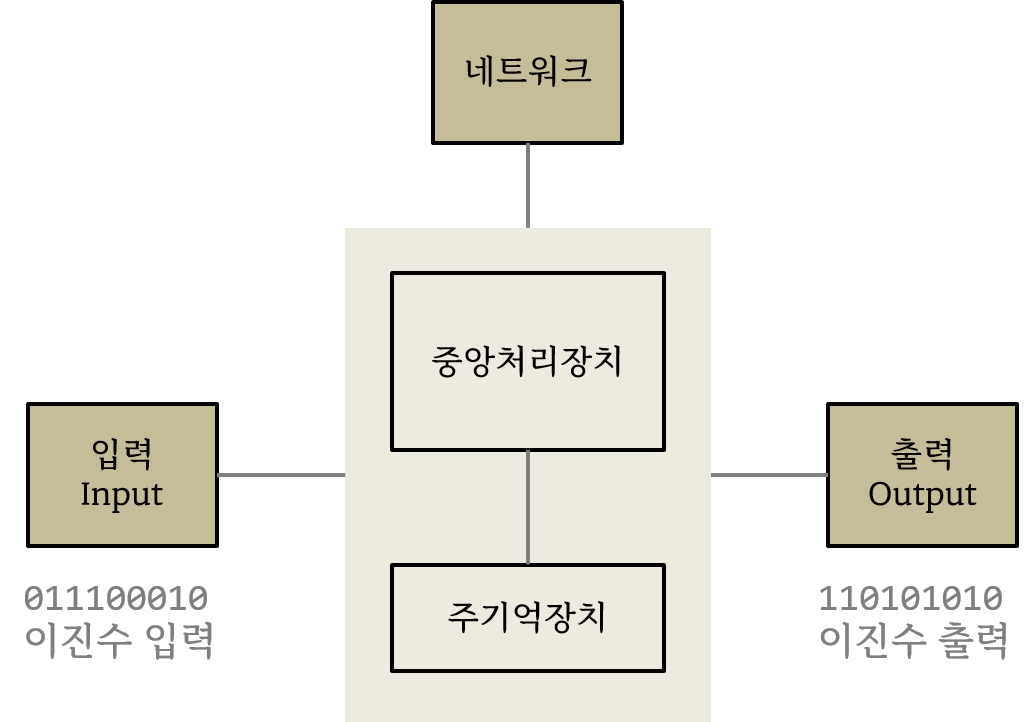
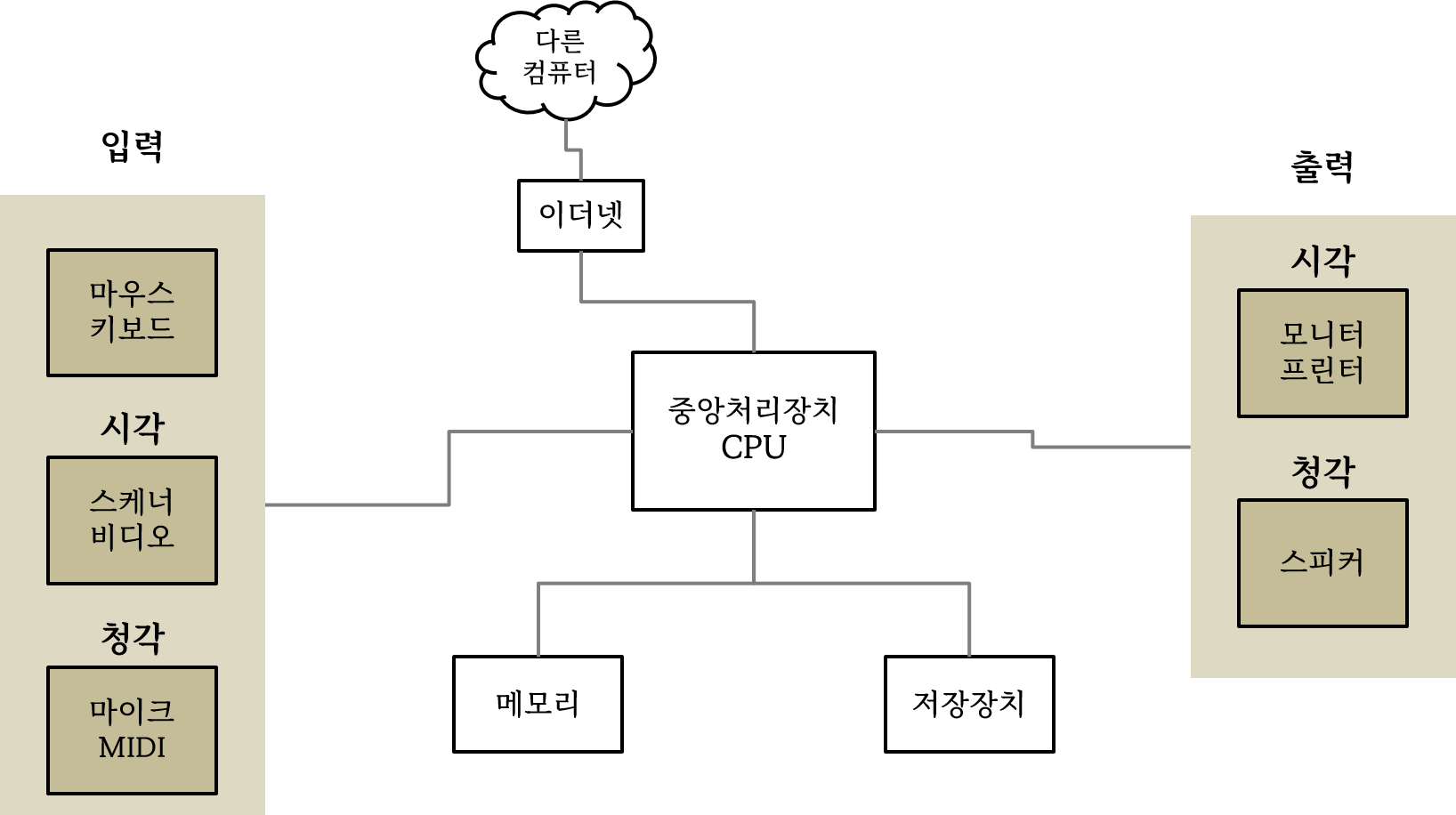
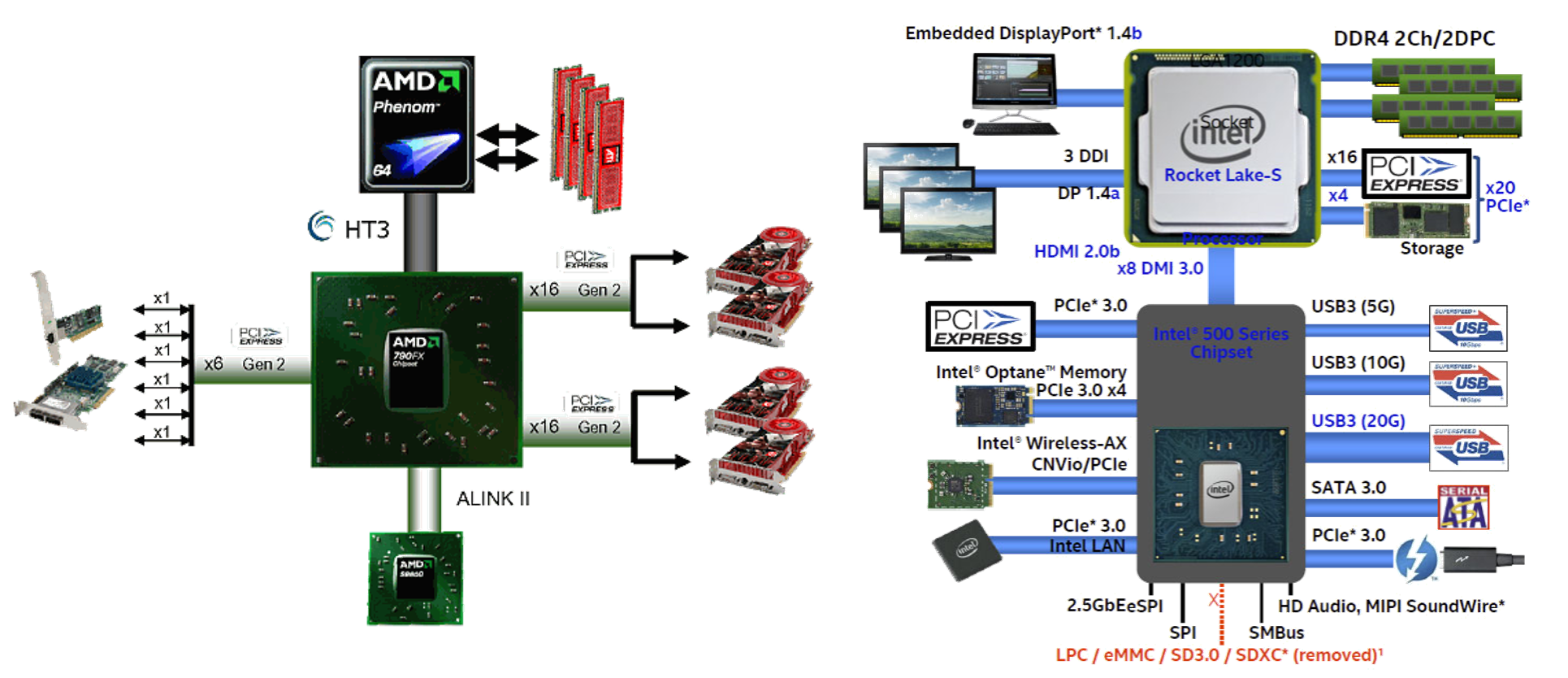
윈텔 아키텍처
과학기술 컴퓨팅
코딩
엔지니어링
복잡성 ↑ → 가치 ↑
Tidyverse API 원칙
-
기존 자료구조를 재사용: Reuse existing data structures.
-
파이프 연산자로 간단한 함수를 조합: Compose simple functions with the pipe.
-
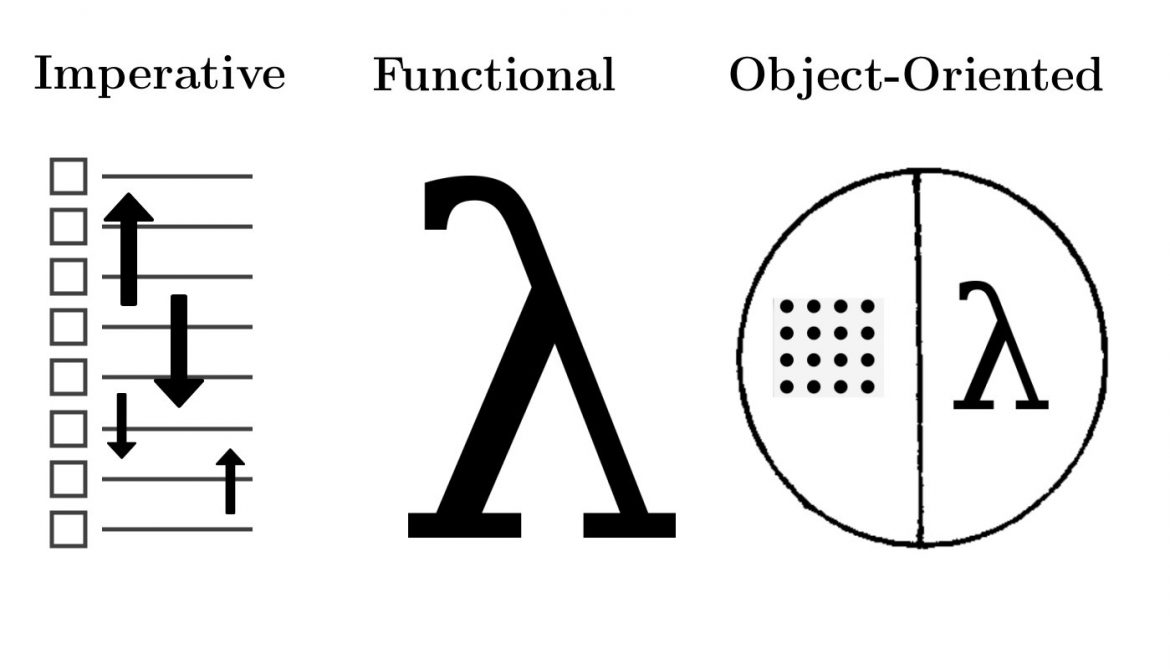
함수형 프로그래밍을 적극 사용: Embrace functional programming.
-
기계가 아닌 인간을 위한 설계: Design for humans.
설계와 디자인 원칙 (Farley 2020)
-
모듈화: modularity
-
정보 은닉: information hiding
-
관심사 분리: separation of concerns
-
낮은 결합도: loosely coupling
-
높은 응집도: high cohesion
데이터 과학 세상
RStudio → Posit
R
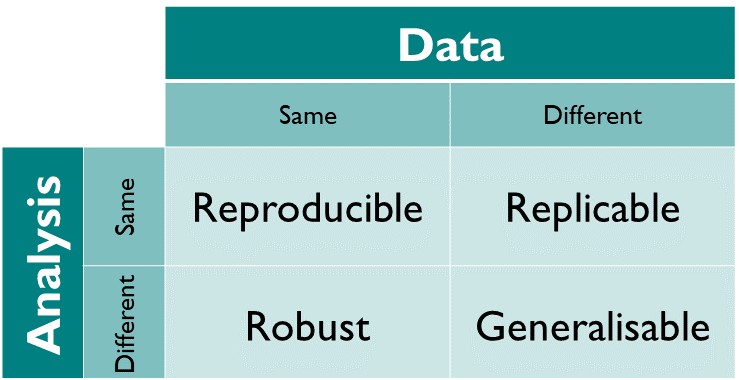
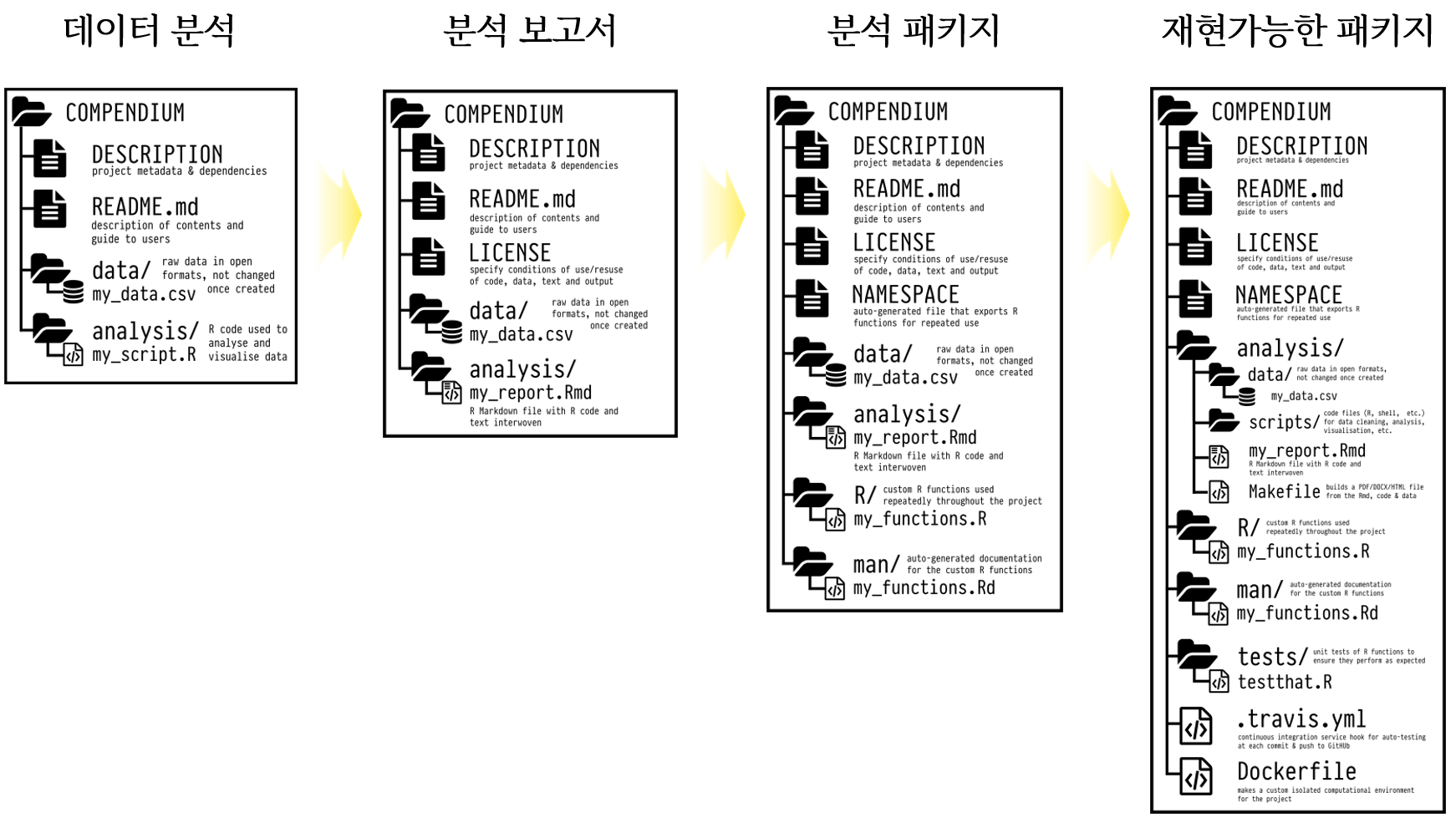
Reproducible: A result is reproducible when the same analysis steps performed on the same dataset consistently produces the same answer.
Replicable: A result is replicable when the same analysis performed on different datasets produces qualitatively similar answers.
Robust: A result is robust when the same dataset is subjected to different analysis workflows to answer the same research question (for example one pipeline written in R and another written in Python) and a qualitatively similar or identical answer is produced. Robust results show that the work is not dependent on the specificities of the programming language chosen to perform the analysis.
Generalisable: Combining replicable and robust findings allow us to form generalisable results. (Community 와/과 Scriberia 2022)
사례
참고문헌