class: center, middle, inverse, title-slide .title[ # 통계학 ] .subtitle[ ## 가설검정 ] .author[ ### 이광춘 ] --- <style type="text/css"> .remark-code{line-height: 1.5; font-size: 30%} </style> --- name: independent_dataset ### 통계적 독립 데이터셋 .panelset[ .panel[.panel-name[R 코드] .pull-left[ ```r library(tidyverse) # Create first dataset dataset1 <- data.frame( value = c(0.72, 0.61, 0.35, 0.19, 0.47, 0.99, 0.12, 0.84, 0.65, 0.38, 0.01, 0.25, 0.96, 0.77, 0.54, 0.82, 0.36, 0.43, 0.06, 0.92, 0.21, 0.17, 0.78, 0.68, 0.94, 0.40, 0.52, 0.29) ) # Create second dataset dataset2 <- data.frame( value = c(0.82, 0.23, 0.67, 0.42, 0.91, 0.17, 0.56, 0.95, 0.03, 0.87, 0.74, 0.45, 0.28, 0.88, 0.51, 0.85, 0.10, 0.72, 0.80, 0.35, 0.57, 0.90, 0.33, 0.06, 0.59, 0.01, 0.23, 0.39) ) ind_df <- bind_cols(dataset1, dataset2) %>% as_tibble() %>% set_names(c("변수1", "변수2")) # ind_df ``` ] .pull-right[ ```r ind_df ``` ``` # A tibble: 28 × 2 변수1 변수2 <dbl> <dbl> 1 0.72 0.82 2 0.61 0.23 3 0.35 0.67 4 0.19 0.42 5 0.47 0.91 6 0.99 0.17 7 0.12 0.56 8 0.84 0.95 9 0.65 0.03 10 0.38 0.87 # … with 18 more rows ``` ] ] .panel[.panel-name[파이썬 코드] ```python # Create first dataset dataset1 = {'value': [0.72, 0.61, 0.35, 0.19, 0.47, 0.99, 0.12, 0.84, 0.65, 0.38, 0.01, 0.25, 0.96, 0.77, 0.54, 0.82, 0.36, 0.43, 0.06, 0.92, 0.21, 0.17, 0.78, 0.68, 0.94, 0.40, 0.52, 0.29]} # Create second dataset dataset2 = {'value': [0.82, 0.23, 0.67, 0.42, 0.91, 0.17, 0.56, 0.95, 0.03, 0.87, 0.74, 0.45, 0.28, 0.88, 0.51, 0.85, 0.10, 0.72, 0.80, 0.35, 0.57, 0.90, 0.33, 0.06, 0.59, 0.01, 0.23, 0.39, 0.64]} # View first few rows of first dataset print(list(dataset1.values())[:5]) # [0.72, 0.61, 0.35, 0.19, 0.47] ``` ] ] --- count: false ##### 통계적으로 독립 : 연속형 두 변수 .panel1-two-independent-continuous-auto[ ```r ## 데이터셋 *ind_df %>% slice_head(n=5) ``` ] .panel2-two-independent-continuous-auto[ ``` # A tibble: 5 × 2 변수1 변수2 <dbl> <dbl> 1 0.72 0.82 2 0.61 0.23 3 0.35 0.67 4 0.19 0.42 5 0.47 0.91 ``` ] --- count: false ##### 통계적으로 독립 : 연속형 두 변수 .panel1-two-independent-continuous-auto[ ```r ## 데이터셋 ind_df %>% slice_head(n=5) # # # ## 시각화 *ggplot(data = ind_df) ``` ] .panel2-two-independent-continuous-auto[ ``` # A tibble: 5 × 2 변수1 변수2 <dbl> <dbl> 1 0.72 0.82 2 0.61 0.23 3 0.35 0.67 4 0.19 0.42 5 0.47 0.91 ``` <!-- --> ] --- count: false ##### 통계적으로 독립 : 연속형 두 변수 .panel1-two-independent-continuous-auto[ ```r ## 데이터셋 ind_df %>% slice_head(n=5) # # # ## 시각화 ggplot(data = ind_df) + * aes(x = 변수1) ``` ] .panel2-two-independent-continuous-auto[ ``` # A tibble: 5 × 2 변수1 변수2 <dbl> <dbl> 1 0.72 0.82 2 0.61 0.23 3 0.35 0.67 4 0.19 0.42 5 0.47 0.91 ``` <!-- --> ] --- count: false ##### 통계적으로 독립 : 연속형 두 변수 .panel1-two-independent-continuous-auto[ ```r ## 데이터셋 ind_df %>% slice_head(n=5) # # # ## 시각화 ggplot(data = ind_df) + aes(x = 변수1) + * aes(y = 변수2) ``` ] .panel2-two-independent-continuous-auto[ ``` # A tibble: 5 × 2 변수1 변수2 <dbl> <dbl> 1 0.72 0.82 2 0.61 0.23 3 0.35 0.67 4 0.19 0.42 5 0.47 0.91 ``` <!-- --> ] --- count: false ##### 통계적으로 독립 : 연속형 두 변수 .panel1-two-independent-continuous-auto[ ```r ## 데이터셋 ind_df %>% slice_head(n=5) # # # ## 시각화 ggplot(data = ind_df) + aes(x = 변수1) + aes(y = 변수2) + * geom_point(color = "steelblue", * alpha = .8) ``` ] .panel2-two-independent-continuous-auto[ ``` # A tibble: 5 × 2 변수1 변수2 <dbl> <dbl> 1 0.72 0.82 2 0.61 0.23 3 0.35 0.67 4 0.19 0.42 5 0.47 0.91 ``` 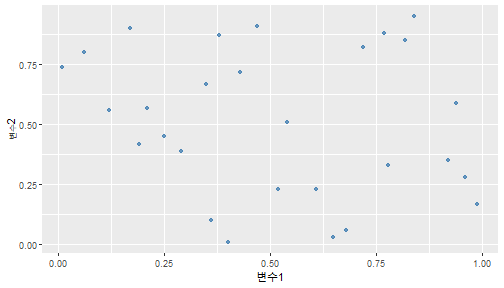<!-- --> ] --- count: false ##### 통계적으로 독립 : 연속형 두 변수 .panel1-two-independent-continuous-auto[ ```r ## 데이터셋 ind_df %>% slice_head(n=5) # # # ## 시각화 ggplot(data = ind_df) + aes(x = 변수1) + aes(y = 변수2) + geom_point(color = "steelblue", alpha = .8) # # # # # # ## 통계검정 *cor.test(x = ind_df$변수1, * y = ind_df$변수1) ``` ] .panel2-two-independent-continuous-auto[ ``` # A tibble: 5 × 2 변수1 변수2 <dbl> <dbl> 1 0.72 0.82 2 0.61 0.23 3 0.35 0.67 4 0.19 0.42 5 0.47 0.91 ``` 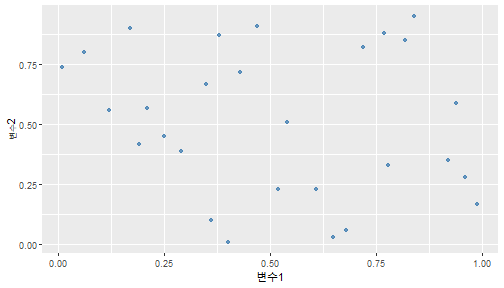<!-- --> ``` Pearson's product-moment correlation data: ind_df$변수1 and ind_df$변수1 t = Inf, df = 26, p-value < 2.2e-16 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: 1 1 sample estimates: cor 1 ``` ] <style> .panel1-two-independent-continuous-auto { color: black; width: 34.3%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-two-independent-continuous-auto { color: black; width: 63.7%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-two-independent-continuous-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- class: center, middle, inverse # Y (연속형), X (연속형) --- ### 통계적으로 독립 : 연속형 두 변수 --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 0.126 -0.817 2 0.838 -2.05 3 -0.517 0.247 ``` 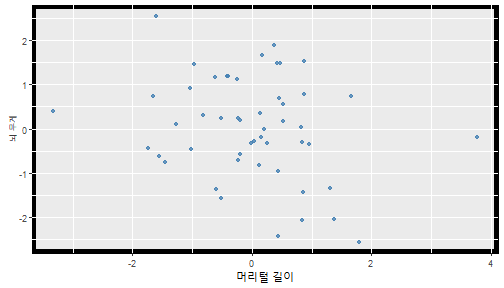<!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = -1.7116, df = 48, p-value = 0.09343 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.48575406 0.04126876 sample estimates: cor -0.2398339 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 -0.992 0.456 2 0.519 -0.982 3 0.544 1.00 ``` 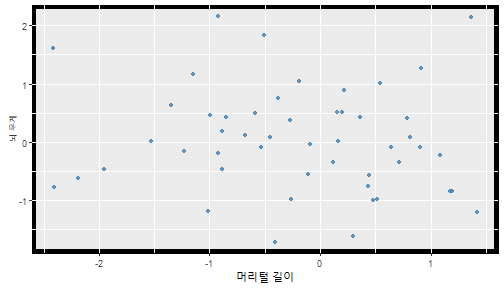<!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = -0.68328, df = 48, p-value = 0.4977 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.3664821 0.1852626 sample estimates: cor -0.09814626 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 0.780 -0.927 2 -0.153 -0.618 3 1.01 -0.690 ``` 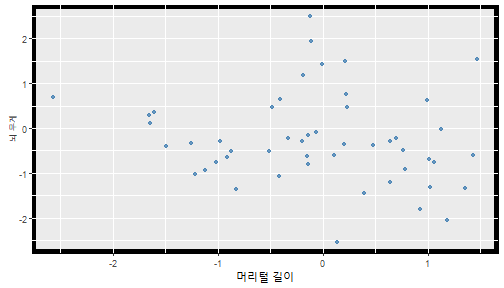<!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = -1.083, df = 48, p-value = 0.2842 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.4149515 0.1294715 sample estimates: cor -0.154442 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 0.760 0.00493 2 0.435 1.46 3 0.799 -1.06 ``` 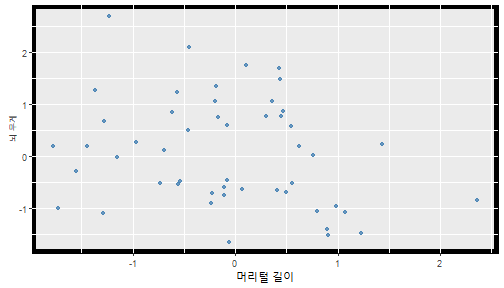<!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = -1.7686, df = 48, p-value = 0.08332 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.49182941 0.03329733 sample estimates: cor -0.2473428 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 2.54 -2.16 2 1.94 -0.502 3 0.344 -0.577 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = 0.91172, df = 48, p-value = 0.3665 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.1534495 0.3944922 sample estimates: cor 0.1304709 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 -1.17 -0.204 2 -0.361 -0.448 3 -1.13 -0.756 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = 1.0593, df = 48, p-value = 0.2948 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.1327916 0.4121511 sample estimates: cor 0.1511427 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 -0.898 -0.875 2 0.703 -0.926 3 0.775 -1.69 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = -0.22933, df = 48, p-value = 0.8196 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.3085887 0.2475446 sample estimates: cor -0.03308263 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 -0.966 0.350 2 -0.0761 0.980 3 -0.547 -0.0916 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = -1.1484, df = 48, p-value = 0.2565 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.4226367 0.1202971 sample estimates: cor -0.1635262 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 -0.689 2.17 2 -1.53 0.675 3 -0.193 1.22 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = 0.98493, df = 48, p-value = 0.3296 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.1432103 0.4032958 sample estimates: cor 0.1407479 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 0.441 0.453 2 -0.283 -0.655 3 -0.372 0.203 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = 0.26905, df = 48, p-value = 0.789 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.2421581 0.3137639 sample estimates: cor 0.03880529 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 0.289 1.25 2 0.0924 1.49 3 1.41 0.669 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = -0.20304, df = 48, p-value = 0.84 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.3051537 0.2511010 sample estimates: cor -0.02929417 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 0.859 -0.719 2 -1.04 -0.569 3 0.826 0.644 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = 0.18744, df = 48, p-value = 0.8521 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.2532097 0.3031101 sample estimates: cor 0.02704411 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 -0.906 -1.47 2 -1.44 -0.600 3 0.820 0.736 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = 1.2368, df = 48, p-value = 0.2222 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.1078909 0.4329063 sample estimates: cor 0.1757343 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 1.38 0.114 2 0.594 -1.77 3 1.83 0.784 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = -0.31639, df = 48, p-value = 0.7531 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.3199051 0.2357213 sample estimates: cor -0.04561959 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 -0.257 0.254 2 2.11 0.197 3 1.51 -0.105 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = -0.36502, df = 48, p-value = 0.7167 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.3261838 0.2290897 sample estimates: cor -0.05261299 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 0.354 0.842 2 -0.134 0.329 3 0.787 0.172 ``` <!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = 0.48954, df = 48, p-value = 0.6267 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.2120241 0.3421190 sample estimates: cor 0.07048327 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 0.623 0.823 2 0.380 0.719 3 -0.197 -1.63 ``` 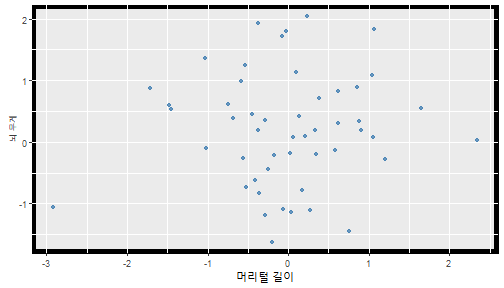<!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = 0.44066, df = 48, p-value = 0.6614 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.2187364 0.3358892 sample estimates: cor 0.06347607 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 2.81 1.36 2 -0.632 2.27 3 0.223 0.496 ``` 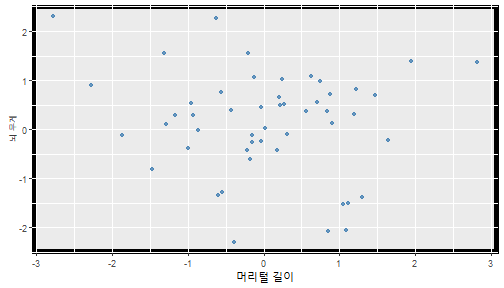<!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = -0.86615, df = 48, p-value = 0.3907 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.3889695 0.1598132 sample estimates: cor -0.1240528 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 0.0784 -0.519 2 0.816 -1.74 3 -0.605 1.66 ``` 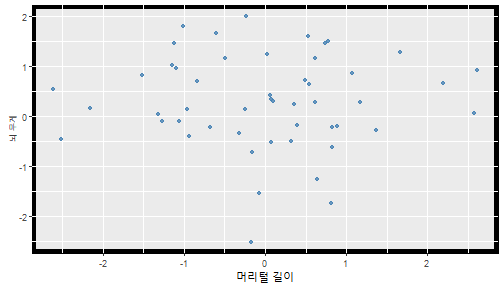<!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = -0.038981, df = 48, p-value = 0.9691 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.2835300 0.2731491 sample estimates: cor -0.00562635 ``` ] --- count: false ##### 두변수(머리털 길이와 뇌 무게) 통계적 독립 .panel1-independent-20[ ```r library(tidyverse) tibble(hair_length = rnorm(n = 50), brain_weight = rnorm(n = 50)) -> ind_data; ind_data %>% slice_head( n = 3) ggplot(data = ind_data) + aes(x = hair_length) + aes(y = brain_weight) + geom_point(color = "steelblue", alpha = .8) + theme(panel.background = element_rect( color = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, "red", "black"), size = ifelse(cor.test(ind_data$hair_length, ind_data$brain_weight)[[3]]<.05, 8, 3))) + labs(x = "머리털 길이", y = "뇌 무게") cor.test(x = ind_data$hair_length, y = ind_data$brain_weight) ``` ] .panel2-independent-20[ ``` # A tibble: 3 × 2 hair_length brain_weight <dbl> <dbl> 1 0.401 0.562 2 0.202 1.74 3 -2.04 0.889 ``` 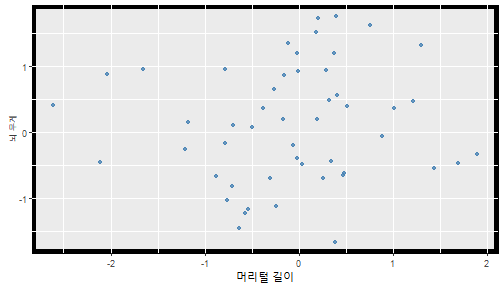<!-- --> ``` Pearson's product-moment correlation data: ind_data$hair_length and ind_data$brain_weight t = 0.54774, df = 48, p-value = 0.5864 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: -0.2040093 0.3494946 sample estimates: cor 0.07881394 ``` ] <style> .panel1-independent-20 { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-independent-20 { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-independent-20 { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- class: center, middle, inverse # Y (연속형), X (2 범주) --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 남성 167. 2 여성 161. 3 여성 183. ``` 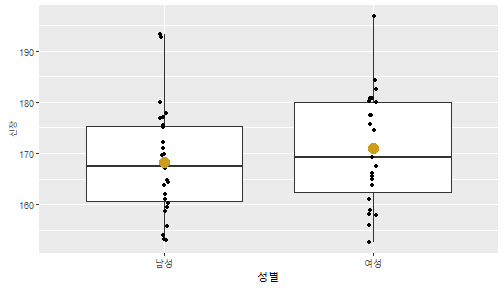<!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -0.85102, df = 45.834, p-value = 0.3992 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -8.855583 3.593026 sample estimates: mean in group 남성 mean in group 여성 168.3132 170.9445 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 여성 177. 2 남성 172. 3 여성 174. ``` 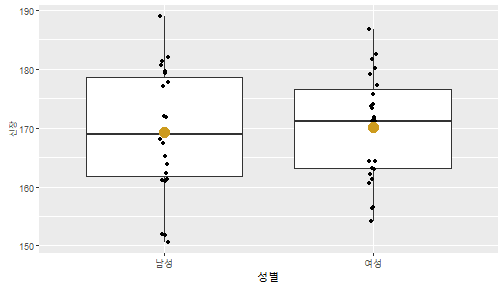<!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -0.3146, df = 42.926, p-value = 0.7546 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -6.444135 4.704963 sample estimates: mean in group 남성 mean in group 여성 169.2713 170.1408 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 여성 187. 2 여성 188. 3 여성 182. ``` 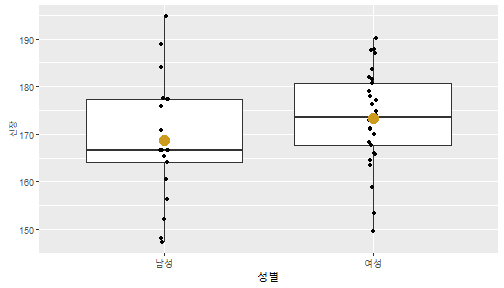<!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -1.4062, df = 37.611, p-value = 0.1679 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -11.18836 2.01795 sample estimates: mean in group 남성 mean in group 여성 168.7906 173.3758 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 남성 172. 2 남성 172. 3 여성 190. ``` 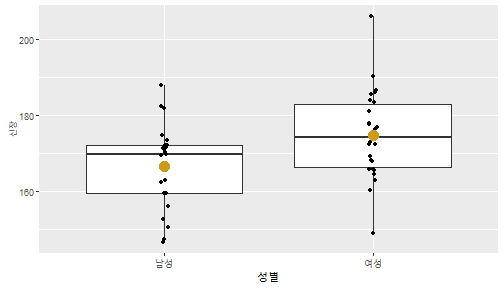<!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -2.5724, df = 47.992, p-value = 0.01325 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -14.464167 -1.772919 sample estimates: mean in group 남성 mean in group 여성 166.6604 174.7789 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 남성 170. 2 여성 172. 3 남성 169. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -0.41433, df = 47.411, p-value = 0.6805 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -7.128098 4.692925 sample estimates: mean in group 남성 mean in group 여성 169.9255 171.1431 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 남성 191. 2 여성 176. 3 여성 184. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -0.56102, df = 42.725, p-value = 0.5777 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -8.507627 4.804919 sample estimates: mean in group 남성 mean in group 여성 168.3479 170.1993 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 여성 172. 2 남성 174. 3 남성 179. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -1.1719, df = 46.689, p-value = 0.2472 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -9.231282 2.435958 sample estimates: mean in group 남성 mean in group 여성 171.0713 174.4690 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 여성 155. 2 남성 163. 3 여성 173. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = 0.12071, df = 45.568, p-value = 0.9045 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -5.033867 5.675951 sample estimates: mean in group 남성 mean in group 여성 168.9828 168.6618 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 남성 160. 2 여성 151. 3 남성 159. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -0.58033, df = 46.956, p-value = 0.5645 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -6.934225 3.829343 sample estimates: mean in group 남성 mean in group 여성 168.2185 169.7710 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 여성 178. 2 남성 175. 3 여성 176. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -1.6925, df = 41.7, p-value = 0.09802 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -8.2292225 0.7230509 sample estimates: mean in group 남성 mean in group 여성 166.4234 170.1765 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 여성 169. 2 남성 184. 3 남성 170. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = 0.82257, df = 47.402, p-value = 0.4149 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -2.931505 6.988623 sample estimates: mean in group 남성 mean in group 여성 174.1101 172.0815 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 남성 147. 2 남성 140. 3 여성 154. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -0.63894, df = 45.309, p-value = 0.5261 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -9.201881 4.769010 sample estimates: mean in group 남성 mean in group 여성 167.5944 169.8109 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 여성 164. 2 남성 160. 3 남성 164. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = 1.5526, df = 47.974, p-value = 0.1271 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -1.225737 9.534091 sample estimates: mean in group 남성 mean in group 여성 172.1478 167.9936 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 여성 172. 2 남성 169. 3 여성 160. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = 1.4574, df = 44.749, p-value = 0.152 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -1.511812 9.423371 sample estimates: mean in group 남성 mean in group 여성 168.9510 164.9952 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 남성 159. 2 남성 175. 3 여성 175. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = 1.7418, df = 47.964, p-value = 0.08795 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -0.6722814 9.3823483 sample estimates: mean in group 남성 mean in group 여성 171.2503 166.8953 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 남성 145. 2 남성 166. 3 여성 170. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -0.6264, df = 47.952, p-value = 0.534 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -6.860411 3.601249 sample estimates: mean in group 남성 mean in group 여성 167.1057 168.7353 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 여성 167. 2 여성 174. 3 남성 186. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = 0.79931, df = 45.599, p-value = 0.4283 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -3.016733 6.988982 sample estimates: mean in group 남성 mean in group 여성 170.3059 168.3197 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 여성 160. 2 여성 164. 3 여성 177. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -0.79109, df = 40.857, p-value = 0.4335 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -7.051308 3.082229 sample estimates: mean in group 남성 mean in group 여성 169.0426 171.0271 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 남성 168. 2 여성 154. 3 남성 183. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = 0.45348, df = 25.427, p-value = 0.654 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -5.290816 8.281870 sample estimates: mean in group 남성 mean in group 여성 167.7157 166.2202 ``` ] --- count: false ##### 두집단(남/녀) 신장이 독립 .panel1-c_and_d2-20[ ```r tibble(sex = sample(x = c("남성","여성"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # # # # visualization ggplot(height_data) + aes(x = sex) + aes(y = height) + aes(group = sex) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs(x= "성별", y = "신장") # statistical test t.test(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$sex) # the discrete variable ``` ] .panel2-c_and_d2-20[ ``` # A tibble: 3 × 2 sex height <chr> <dbl> 1 여성 170. 2 여성 170. 3 남성 183. ``` <!-- --> ``` Welch Two Sample t-test data: height_data$height by height_data$sex t = -0.1438, df = 46.298, p-value = 0.8863 alternative hypothesis: true difference in means between group 남성 and group 여성 is not equal to 0 95 percent confidence interval: -5.773838 5.003770 sample estimates: mean in group 남성 mean in group 여성 172.4989 172.8839 ``` ] <style> .panel1-c_and_d2-20 { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-c_and_d2-20 { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-c_and_d2-20 { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- class: center, middle, inverse # Y (연속형), X (N 범주) --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 1반 173. 2 3반 170. 3 2반 205. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -0.507663765 -12.875113 11.859785 0.9995242 3반-1반 -2.954968492 -13.171057 7.261120 0.8671120 특수반-1반 -0.505066084 -11.352036 10.341904 0.9993065 3반-2반 -2.447304727 -14.064500 9.169891 0.9429070 특수반-2반 0.002597681 -12.173096 12.178292 1.0000000 특수반-3반 2.449902408 -7.533193 12.432998 0.9135634 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 1반 172. 2 3반 172. 3 1반 162. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 0.9415869 -11.19792 13.081099 0.9968290 3반-1반 -1.0328743 -12.98240 10.916649 0.9956295 특수반-1반 -3.0043967 -15.14391 9.135115 0.9115892 3반-2반 -1.9744613 -13.09063 9.141709 0.9645455 특수반-2반 -3.9459837 -15.26614 7.374170 0.7894170 특수반-3반 -1.9715224 -13.08769 9.144647 0.9646944 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 1반 167. 2 3반 171. 3 2반 167. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -1.5178444 -11.721244 8.685556 0.9786196 3반-1반 -0.7390442 -9.575448 8.097359 0.9960354 특수반-1반 -4.3012202 -13.907404 5.304963 0.6340056 3반-2반 0.7788002 -9.424600 10.982200 0.9969766 특수반-2반 -2.7833758 -13.660237 8.093485 0.9033799 특수반-3반 -3.5621760 -13.168360 6.044008 0.7567074 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 2반 160. 2 1반 166. 3 특수반 155. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -0.8917641 -13.07714 11.29361 0.9973315 3반-1반 0.1908132 -12.28924 12.67087 0.9999753 특수반-1반 -1.6278026 -13.55815 10.30254 0.9833398 3반-2반 1.0825773 -11.84699 14.01214 0.9960224 특수반-2반 -0.7360386 -13.13584 11.66376 0.9985687 특수반-3반 -1.8186159 -14.50812 10.87088 0.9807962 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 1반 171. 2 1반 177. 3 3반 157. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -7.029198 -17.683519 3.6251236 0.3061771 3반-1반 -8.690892 -17.745324 0.3635402 0.0641399 특수반-1반 -2.326404 -11.380836 6.7280286 0.9023413 3반-2반 -1.661694 -11.387714 8.0643263 0.9682400 특수반-2반 4.702794 -5.023226 14.4288146 0.5745900 특수반-3반 6.364488 -1.576774 14.3057506 0.1570043 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 2반 183. 2 3반 186. 3 1반 163. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 6.0228745 -5.284201 17.329950 0.4937312 3반-1반 4.9100396 -4.945968 14.766048 0.5502779 특수반-1반 6.9684918 -4.699264 18.636247 0.3931763 3반-2반 -1.1128349 -11.996273 9.770603 0.9928257 특수반-2반 0.9456173 -11.602085 13.493320 0.9970887 특수반-3반 2.0584522 -9.199247 13.316151 0.9615234 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 특수반 173. 2 3반 171. 3 1반 160. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 0.1944076 -10.008963 10.397778 0.9999522 3반-1반 -3.0937827 -12.288317 6.100751 0.8064722 특수반-1반 3.5734003 -5.276646 12.423447 0.7055770 3반-2반 -3.2881903 -12.776018 6.199637 0.7922900 특수반-2반 3.3789927 -5.775390 12.533376 0.7592784 특수반-3반 6.6671831 -1.347392 14.681758 0.1337197 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 1반 178. 2 2반 181. 3 특수반 157. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -0.6269522 -12.068942 10.815038 0.9988725 3반-1반 -5.9847571 -17.838865 5.869351 0.5392161 특수반-1반 -2.4449258 -14.078938 9.189086 0.9432865 3반-2반 -5.3578049 -16.529625 5.814016 0.5811496 특수반-2반 -1.8179736 -12.755978 9.120031 0.9706323 특수반-3반 3.5398313 -7.828576 14.908238 0.8399599 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 3반 170. 2 1반 159. 3 특수반 151. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 7.362096 -4.288616 19.012807 0.3434756 3반-1반 5.310581 -6.930985 17.552148 0.6569685 특수반-1반 1.685360 -9.342887 12.713606 0.9768984 3반-2반 -2.051514 -14.512875 10.409846 0.9714190 특수반-2반 -5.676736 -16.948461 5.594989 0.5412664 특수반-3반 -3.625222 -15.506663 8.256219 0.8478744 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 1반 162. 2 특수반 169. 3 특수반 159. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 4.3819317 -6.511688 15.27555 0.7080003 3반-1반 3.6282720 -7.265348 14.52189 0.8111850 특수반-1반 4.2143404 -6.679279 15.10796 0.7321629 3반-2반 -0.7536597 -12.058503 10.55118 0.9979775 특수반-2반 -0.1675913 -11.472435 11.13725 0.9999775 특수반-3반 0.5860684 -10.718775 11.89091 0.9990442 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 2반 182. 2 3반 196. 3 특수반 185. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 4.9569108 -6.345439 16.259260 0.6491523 3반-1반 8.4427600 -3.806325 20.691845 0.2694392 특수반-1반 0.6507311 -11.032703 12.334165 0.9988162 3반-2반 3.4858492 -6.714767 13.686465 0.7991426 특수반-2반 -4.3061796 -13.820123 5.207763 0.6259570 특수반-3반 -7.7920288 -18.413332 2.829275 0.2198131 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 2반 167. 2 3반 169. 3 1반 165. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -1.2784275 -9.098871 6.542016 0.9719880 3반-1반 -0.9964659 -9.306151 7.313219 0.9885548 특수반-1반 0.7856471 -9.075129 10.646423 0.9965663 3반-2반 0.2819616 -8.145597 8.709521 0.9997419 특수반-2반 2.0640746 -7.896236 12.024385 0.9454379 특수반-3반 1.7821130 -8.566767 12.130992 0.9675213 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 특수반 176. 2 특수반 183. 3 특수반 182. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 2.608015 -8.460510 13.676540 0.9225075 3반-1반 3.440079 -8.168688 15.048846 0.8586704 특수반-1반 1.416289 -8.937368 11.769946 0.9832161 3반-2반 0.832064 -10.776703 12.440831 0.9974915 특수반-2반 -1.191726 -11.545383 9.161931 0.9898464 특수반-3반 -2.023790 -12.953082 8.905503 0.9601327 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 1반 184. 2 3반 159. 3 2반 173. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -8.611609 -20.191754 2.968535 0.2095387 3반-1반 -3.545602 -15.713022 8.621819 0.8645720 특수반-1반 -2.492975 -13.454424 8.468474 0.9295892 3반-2반 5.066008 -7.319876 17.451891 0.6972066 특수반-2반 6.118634 -5.084819 17.322087 0.4720622 특수반-3반 1.052627 -10.756850 12.862103 0.9952138 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 1반 181. 2 특수반 155. 3 3반 148. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -0.6167487 -14.213979 12.98048 0.9993587 3반-1반 -3.1711394 -16.382554 10.04027 0.9185444 특수반-1반 1.2636609 -11.331134 13.85846 0.9932146 3반-2반 -2.5543907 -17.292340 12.18356 0.9669205 특수반-2반 1.8804096 -12.307421 16.06824 0.9846849 특수반-3반 4.4348003 -9.383714 18.25332 0.8275308 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 특수반 181. 2 3반 174. 3 특수반 170. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -1.7022876 -12.013301 8.608726 0.9711888 3반-1반 -0.6117371 -9.147579 7.924105 0.9974924 특수반-1반 1.5169569 -6.708386 9.742300 0.9605828 3반-2반 1.0905505 -9.469816 11.650917 0.9926137 특수반-2반 3.2192446 -7.091769 13.530258 0.8388842 특수반-3반 2.1286941 -6.407148 10.664536 0.9097742 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 3반 187. 2 1반 156. 3 2반 183. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -1.710227 -12.856377 9.435924 0.9766309 3반-1반 2.187413 -7.946802 12.321628 0.9389604 특수반-1반 0.836745 -12.216290 13.889780 0.9982003 3반-2반 3.897639 -6.478862 14.274141 0.7494392 특수반-2반 2.546972 -10.695052 15.788995 0.9556536 특수반-3반 -1.350668 -13.752959 11.051624 0.9913661 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 1반 153. 2 3반 161. 3 2반 161. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 2.334310 -10.908206 15.576826 0.9653001 3반-1반 4.332345 -9.044616 17.709306 0.8236343 특수반-1반 2.176488 -11.066028 15.419004 0.9715543 3반-2반 1.998035 -8.189556 12.185625 0.9531745 특수반-2반 -0.157822 -10.168223 9.852579 0.9999729 특수반-3반 -2.155857 -12.343447 8.031734 0.9421920 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 1반 163. 2 1반 166. 3 3반 174. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -2.2323418 -11.552461 7.087777 0.9190102 3반-1반 7.0739378 -3.165037 17.312913 0.2674992 특수반-1반 -0.9556717 -10.911486 9.000143 0.9940434 3반-2반 9.3062796 -1.077936 19.690495 0.0935213 특수반-2반 1.2766701 -8.828456 11.381796 0.9866748 특수반-3반 -8.0296095 -18.987965 2.928746 0.2207354 ``` ] --- count: false ##### N 학급 신장이 독립 .panel1-c_and_d3-20[ ```r tibble(class = sample(x = c("1반","2반","3반","특수반"), size = 50, replace = TRUE)) %>% mutate(height = rnorm(n = 50, mean = 170, sd = 10)) -> height_data; height_data %>% slice_head( n = 3) # # # # # visualization ggplot(height_data) + aes(x = class) + aes(y = height) + aes(group = class) + geom_boxplot() + geom_jitter(height = 0, width = .02) + stat_summary(fun.y = mean, geom = "point", col = "goldenrod3", size = 5) + labs( x = "학년", y = "신장") # statistical test TukeyHSD(aov(height_data$height ~ # the continuous variable by (~) # the continuous variable by (~) height_data$class)) # the discrete variable ``` ] .panel2-c_and_d3-20[ ``` # A tibble: 3 × 2 class height <chr> <dbl> 1 1반 164. 2 특수반 166. 3 2반 154. ``` <!-- --> ``` Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = height_data$height ~ height_data$class) $`height_data$class` diff lwr upr p adj 2반-1반 -1.4028727 -13.520144 10.71440 0.9896719 3반-1반 -0.3325582 -12.088038 11.42292 0.9998439 특수반-1반 0.1860896 -12.263223 12.63540 0.9999769 3반-2반 1.0703145 -8.376831 10.51746 0.9903059 특수반-2반 1.5889623 -8.708738 11.88666 0.9762505 특수반-3반 0.5186478 -9.350784 10.38808 0.9990047 ``` ] <style> .panel1-c_and_d3-20 { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-c_and_d3-20 { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-c_and_d3-20 { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- class: center, middle, inverse # Y (범주), X (범주) --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 남자 오른손잡이 2 남자 오른손잡이 3 남자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 40 14 여자 32 14 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.076761, df = 1, p-value = 0.7817 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 오른손잡이 2 남자 오른손잡이 3 남자 왼손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 31 8 여자 42 19 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.87884, df = 1, p-value = 0.3485 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 남자 왼손잡이 2 여자 오른손잡이 3 여자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 29 24 여자 34 13 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 2.6061, df = 1, p-value = 0.1065 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 오른손잡이 2 남자 오른손잡이 3 여자 왼손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 38 14 여자 29 19 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 1.2821, df = 1, p-value = 0.2575 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 남자 왼손잡이 2 남자 오른손잡이 3 여자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 34 20 여자 38 8 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 3.8309, df = 1, p-value = 0.05031 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 오른손잡이 2 남자 오른손잡이 3 남자 왼손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 37 16 여자 32 15 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0, df = 1, p-value = 1 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 오른손잡이 2 여자 왼손잡이 3 여자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 37 14 여자 33 16 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.12195, df = 1, p-value = 0.7269 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 오른손잡이 2 여자 오른손잡이 3 남자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 35 15 여자 37 13 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.049603, df = 1, p-value = 0.8238 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 오른손잡이 2 여자 오른손잡이 3 남자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 35 15 여자 34 16 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0, df = 1, p-value = 1 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 왼손잡이 2 여자 오른손잡이 3 여자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 38 11 여자 44 7 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.76518, df = 1, p-value = 0.3817 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 오른손잡이 2 여자 왼손잡이 3 남자 왼손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 39 14 여자 33 14 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.023019, df = 1, p-value = 0.8794 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 오른손잡이 2 남자 오른손잡이 3 여자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 39 13 여자 31 17 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.84135, df = 1, p-value = 0.359 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 왼손잡이 2 여자 오른손잡이 3 여자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 38 14 여자 32 16 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.23085, df = 1, p-value = 0.6309 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 남자 오른손잡이 2 남자 왼손잡이 3 남자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 32 16 여자 33 19 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.01585, df = 1, p-value = 0.8998 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 남자 오른손잡이 2 남자 오른손잡이 3 남자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 41 12 여자 34 13 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.12043, df = 1, p-value = 0.7286 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 오른손잡이 2 남자 왼손잡이 3 여자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 32 20 여자 34 14 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.59139, df = 1, p-value = 0.4419 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 남자 오른손잡이 2 남자 오른손잡이 3 남자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 39 14 여자 35 12 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 4.8721e-31, df = 1, p-value = 1 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 남자 왼손잡이 2 남자 오른손잡이 3 여자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 32 16 여자 37 15 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.071999, df = 1, p-value = 0.7884 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 남자 왼손잡이 2 남자 오른손잡이 3 남자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 36 12 여자 43 9 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.48695, df = 1, p-value = 0.4853 ``` ] --- count: false .panel1-d_and_d-20[ ```r # 데이터 tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = TRUE, prob = c(.3,.7))) -> handed_data ; handed_data %>% slice_head( n = 3) # 시각화 library(ggmosaic); library(janitor); library(patchwork) ggplot(data = handed_data) + geom_mosaic(aes(x = product(sex), fill = handed)) + labs(x = "성별", y = "손잡이", fill = "손잡이 구분") # 표 table(x = handed_data$sex, y = handed_data$handed) # # # # # 검정통계량 chisq.test(x = handed_data$sex, y = handed_data$handed) ``` ] .panel2-d_and_d-20[ ``` # A tibble: 3 × 2 sex handed <chr> <chr> 1 여자 오른손잡이 2 여자 왼손잡이 3 남자 오른손잡이 ``` <!-- --> ``` y x 오른손잡이 왼손잡이 남자 33 19 여자 32 16 ``` ``` Pearson's Chi-squared test with Yates' continuity correction data: handed_data$sex and handed_data$handed X-squared = 0.01585, df = 1, p-value = 0.8998 ``` ] <style> .panel1-d_and_d-20 { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-d_and_d-20 { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-d_and_d-20 { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r *library(gt); library(janitor) ``` ] .panel2-gt_table-auto[ ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) *handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), * handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) ``` ] .panel2-gt_table-auto[ ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 *handed_data ``` ] .panel2-gt_table-auto[ ``` # A tibble: 100 × 2 sex handed <chr> <chr> 1 여자 왼손잡이 2 남자 오른손잡이 3 남자 오른손잡이 4 남자 오른손잡이 5 남자 오른손잡이 6 여자 왼손잡이 7 남자 오른손잡이 8 남자 오른손잡이 9 여자 오른손잡이 10 남자 오른손잡이 # … with 90 more rows ``` ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% * tabyl(sex, handed) ``` ] .panel2-gt_table-auto[ ``` sex 오른손잡이 왼손잡이 남자 33 11 여자 36 20 ``` ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% tabyl(sex, handed) %>% * adorn_totals(where = c("row", "col"), name = "합계") ``` ] .panel2-gt_table-auto[ ``` sex 오른손잡이 왼손잡이 합계 남자 45 11 56 여자 28 16 44 합계 73 27 100 ``` ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% tabyl(sex, handed) %>% adorn_totals(where = c("row", "col"), name = "합계") %>% * adorn_percentages(denominator = "col") ``` ] .panel2-gt_table-auto[ ``` sex 오른손잡이 왼손잡이 합계 남자 0.5822785 0.3809524 0.54 여자 0.4177215 0.6190476 0.46 합계 1.0000000 1.0000000 1.00 ``` ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% tabyl(sex, handed) %>% adorn_totals(where = c("row", "col"), name = "합계") %>% adorn_percentages(denominator = "col") %>% * adorn_pct_formatting(digits = 0) ``` ] .panel2-gt_table-auto[ ``` sex 오른손잡이 왼손잡이 합계 남자 42% 66% 49% 여자 58% 34% 51% 합계 100% 100% 100% ``` ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% tabyl(sex, handed) %>% adorn_totals(where = c("row", "col"), name = "합계") %>% adorn_percentages(denominator = "col") %>% adorn_pct_formatting(digits = 0) %>% * adorn_ns(position = "rear") ``` ] .panel2-gt_table-auto[ ``` sex 오른손잡이 왼손잡이 합계 남자 60% (40) 52% (17) 57% (57) 여자 40% (27) 48% (16) 43% (43) 합계 100% (67) 100% (33) 100% (100) ``` ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% tabyl(sex, handed) %>% adorn_totals(where = c("row", "col"), name = "합계") %>% adorn_percentages(denominator = "col") %>% adorn_pct_formatting(digits = 0) %>% adorn_ns(position = "rear") %>% # 표 외양 * gt() ``` ] .panel2-gt_table-auto[ <div id="ecgvwswbkz" style="padding-left:0px;padding-right:0px;padding-top:10px;padding-bottom:10px;overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #ecgvwswbkz .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #ecgvwswbkz .gt_heading { background-color: #FFFFFF; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #ecgvwswbkz .gt_caption { padding-top: 4px; padding-bottom: 4px; } #ecgvwswbkz .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #ecgvwswbkz .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; border-top-color: #FFFFFF; border-top-width: 0; } #ecgvwswbkz .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #ecgvwswbkz .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #ecgvwswbkz .gt_col_heading { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #ecgvwswbkz .gt_column_spanner_outer { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #ecgvwswbkz .gt_column_spanner_outer:first-child { padding-left: 0; } #ecgvwswbkz .gt_column_spanner_outer:last-child { padding-right: 0; } #ecgvwswbkz .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #ecgvwswbkz .gt_group_heading { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; text-align: left; } #ecgvwswbkz .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #ecgvwswbkz .gt_from_md > :first-child { margin-top: 0; } #ecgvwswbkz .gt_from_md > :last-child { margin-bottom: 0; } #ecgvwswbkz .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #ecgvwswbkz .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; } #ecgvwswbkz .gt_stub_row_group { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; vertical-align: top; } #ecgvwswbkz .gt_row_group_first td { border-top-width: 2px; } #ecgvwswbkz .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #ecgvwswbkz .gt_first_summary_row { border-top-style: solid; border-top-color: #D3D3D3; } #ecgvwswbkz .gt_first_summary_row.thick { border-top-width: 2px; } #ecgvwswbkz .gt_last_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #ecgvwswbkz .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #ecgvwswbkz .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #ecgvwswbkz .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #ecgvwswbkz .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #ecgvwswbkz .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #ecgvwswbkz .gt_footnote { margin: 0px; font-size: 90%; padding-left: 4px; padding-right: 4px; padding-left: 5px; padding-right: 5px; } #ecgvwswbkz .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #ecgvwswbkz .gt_sourcenote { font-size: 90%; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; } #ecgvwswbkz .gt_left { text-align: left; } #ecgvwswbkz .gt_center { text-align: center; } #ecgvwswbkz .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #ecgvwswbkz .gt_font_normal { font-weight: normal; } #ecgvwswbkz .gt_font_bold { font-weight: bold; } #ecgvwswbkz .gt_font_italic { font-style: italic; } #ecgvwswbkz .gt_super { font-size: 65%; } #ecgvwswbkz .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 75%; vertical-align: 0.4em; } #ecgvwswbkz .gt_asterisk { font-size: 100%; vertical-align: 0; } #ecgvwswbkz .gt_indent_1 { text-indent: 5px; } #ecgvwswbkz .gt_indent_2 { text-indent: 10px; } #ecgvwswbkz .gt_indent_3 { text-indent: 15px; } #ecgvwswbkz .gt_indent_4 { text-indent: 20px; } #ecgvwswbkz .gt_indent_5 { text-indent: 25px; } </style> <table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" scope="col" id="sex">sex</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="오른손잡이">오른손잡이</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="왼손잡이">왼손잡이</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="합계">합계</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td headers="sex" class="gt_row gt_left">남자</td> <td headers="오른손잡이" class="gt_row gt_right">44% (31)</td> <td headers="왼손잡이" class="gt_row gt_right">55% (16)</td> <td headers="합계" class="gt_row gt_right">47% (47)</td></tr> <tr><td headers="sex" class="gt_row gt_left">여자</td> <td headers="오른손잡이" class="gt_row gt_right">56% (40)</td> <td headers="왼손잡이" class="gt_row gt_right">45% (13)</td> <td headers="합계" class="gt_row gt_right">53% (53)</td></tr> <tr><td headers="sex" class="gt_row gt_left">합계</td> <td headers="오른손잡이" class="gt_row gt_right">100% (71)</td> <td headers="왼손잡이" class="gt_row gt_right">100% (29)</td> <td headers="합계" class="gt_row gt_right">100% (100)</td></tr> </tbody> </table> </div> ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% tabyl(sex, handed) %>% adorn_totals(where = c("row", "col"), name = "합계") %>% adorn_percentages(denominator = "col") %>% adorn_pct_formatting(digits = 0) %>% adorn_ns(position = "rear") %>% # 표 외양 gt() %>% * tab_header( * title = md("**☛ 2변량 교차 범주형 데이터 ☚**"), * ) ``` ] .panel2-gt_table-auto[ <div id="yeancciakz" style="padding-left:0px;padding-right:0px;padding-top:10px;padding-bottom:10px;overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #yeancciakz .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #yeancciakz .gt_heading { background-color: #FFFFFF; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #yeancciakz .gt_caption { padding-top: 4px; padding-bottom: 4px; } #yeancciakz .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #yeancciakz .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; border-top-color: #FFFFFF; border-top-width: 0; } #yeancciakz .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #yeancciakz .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #yeancciakz .gt_col_heading { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #yeancciakz .gt_column_spanner_outer { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #yeancciakz .gt_column_spanner_outer:first-child { padding-left: 0; } #yeancciakz .gt_column_spanner_outer:last-child { padding-right: 0; } #yeancciakz .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #yeancciakz .gt_group_heading { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; text-align: left; } #yeancciakz .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #yeancciakz .gt_from_md > :first-child { margin-top: 0; } #yeancciakz .gt_from_md > :last-child { margin-bottom: 0; } #yeancciakz .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #yeancciakz .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; } #yeancciakz .gt_stub_row_group { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; vertical-align: top; } #yeancciakz .gt_row_group_first td { border-top-width: 2px; } #yeancciakz .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #yeancciakz .gt_first_summary_row { border-top-style: solid; border-top-color: #D3D3D3; } #yeancciakz .gt_first_summary_row.thick { border-top-width: 2px; } #yeancciakz .gt_last_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #yeancciakz .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #yeancciakz .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #yeancciakz .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #yeancciakz .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #yeancciakz .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #yeancciakz .gt_footnote { margin: 0px; font-size: 90%; padding-left: 4px; padding-right: 4px; padding-left: 5px; padding-right: 5px; } #yeancciakz .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #yeancciakz .gt_sourcenote { font-size: 90%; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; } #yeancciakz .gt_left { text-align: left; } #yeancciakz .gt_center { text-align: center; } #yeancciakz .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #yeancciakz .gt_font_normal { font-weight: normal; } #yeancciakz .gt_font_bold { font-weight: bold; } #yeancciakz .gt_font_italic { font-style: italic; } #yeancciakz .gt_super { font-size: 65%; } #yeancciakz .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 75%; vertical-align: 0.4em; } #yeancciakz .gt_asterisk { font-size: 100%; vertical-align: 0; } #yeancciakz .gt_indent_1 { text-indent: 5px; } #yeancciakz .gt_indent_2 { text-indent: 10px; } #yeancciakz .gt_indent_3 { text-indent: 15px; } #yeancciakz .gt_indent_4 { text-indent: 20px; } #yeancciakz .gt_indent_5 { text-indent: 25px; } </style> <table class="gt_table"> <thead class="gt_header"> <tr> <td colspan="4" class="gt_heading gt_title gt_font_normal gt_bottom_border" style><strong>☛ 2변량 교차 범주형 데이터 ☚</strong></td> </tr> </thead> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" scope="col" id="sex">sex</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="오른손잡이">오른손잡이</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="왼손잡이">왼손잡이</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="합계">합계</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td headers="sex" class="gt_row gt_left">남자</td> <td headers="오른손잡이" class="gt_row gt_right">42% (30)</td> <td headers="왼손잡이" class="gt_row gt_right">38% (11)</td> <td headers="합계" class="gt_row gt_right">41% (41)</td></tr> <tr><td headers="sex" class="gt_row gt_left">여자</td> <td headers="오른손잡이" class="gt_row gt_right">58% (41)</td> <td headers="왼손잡이" class="gt_row gt_right">62% (18)</td> <td headers="합계" class="gt_row gt_right">59% (59)</td></tr> <tr><td headers="sex" class="gt_row gt_left">합계</td> <td headers="오른손잡이" class="gt_row gt_right">100% (71)</td> <td headers="왼손잡이" class="gt_row gt_right">100% (29)</td> <td headers="합계" class="gt_row gt_right">100% (100)</td></tr> </tbody> </table> </div> ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% tabyl(sex, handed) %>% adorn_totals(where = c("row", "col"), name = "합계") %>% adorn_percentages(denominator = "col") %>% adorn_pct_formatting(digits = 0) %>% adorn_ns(position = "rear") %>% # 표 외양 gt() %>% tab_header( title = md("**☛ 2변량 교차 범주형 데이터 ☚**"), ) %>% * cols_align( * align = "right", * columns = c(오른손잡이, 왼손잡이, 합계) * ) ``` ] .panel2-gt_table-auto[ <div id="uetiojngfk" style="padding-left:0px;padding-right:0px;padding-top:10px;padding-bottom:10px;overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #uetiojngfk .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #uetiojngfk .gt_heading { background-color: #FFFFFF; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #uetiojngfk .gt_caption { padding-top: 4px; padding-bottom: 4px; } #uetiojngfk .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #uetiojngfk .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; border-top-color: #FFFFFF; border-top-width: 0; } #uetiojngfk .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #uetiojngfk .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #uetiojngfk .gt_col_heading { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #uetiojngfk .gt_column_spanner_outer { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #uetiojngfk .gt_column_spanner_outer:first-child { padding-left: 0; } #uetiojngfk .gt_column_spanner_outer:last-child { padding-right: 0; } #uetiojngfk .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #uetiojngfk .gt_group_heading { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; text-align: left; } #uetiojngfk .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #uetiojngfk .gt_from_md > :first-child { margin-top: 0; } #uetiojngfk .gt_from_md > :last-child { margin-bottom: 0; } #uetiojngfk .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #uetiojngfk .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; } #uetiojngfk .gt_stub_row_group { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; vertical-align: top; } #uetiojngfk .gt_row_group_first td { border-top-width: 2px; } #uetiojngfk .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #uetiojngfk .gt_first_summary_row { border-top-style: solid; border-top-color: #D3D3D3; } #uetiojngfk .gt_first_summary_row.thick { border-top-width: 2px; } #uetiojngfk .gt_last_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #uetiojngfk .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #uetiojngfk .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #uetiojngfk .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #uetiojngfk .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #uetiojngfk .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #uetiojngfk .gt_footnote { margin: 0px; font-size: 90%; padding-left: 4px; padding-right: 4px; padding-left: 5px; padding-right: 5px; } #uetiojngfk .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #uetiojngfk .gt_sourcenote { font-size: 90%; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; } #uetiojngfk .gt_left { text-align: left; } #uetiojngfk .gt_center { text-align: center; } #uetiojngfk .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #uetiojngfk .gt_font_normal { font-weight: normal; } #uetiojngfk .gt_font_bold { font-weight: bold; } #uetiojngfk .gt_font_italic { font-style: italic; } #uetiojngfk .gt_super { font-size: 65%; } #uetiojngfk .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 75%; vertical-align: 0.4em; } #uetiojngfk .gt_asterisk { font-size: 100%; vertical-align: 0; } #uetiojngfk .gt_indent_1 { text-indent: 5px; } #uetiojngfk .gt_indent_2 { text-indent: 10px; } #uetiojngfk .gt_indent_3 { text-indent: 15px; } #uetiojngfk .gt_indent_4 { text-indent: 20px; } #uetiojngfk .gt_indent_5 { text-indent: 25px; } </style> <table class="gt_table"> <thead class="gt_header"> <tr> <td colspan="4" class="gt_heading gt_title gt_font_normal gt_bottom_border" style><strong>☛ 2변량 교차 범주형 데이터 ☚</strong></td> </tr> </thead> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" scope="col" id="sex">sex</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="오른손잡이">오른손잡이</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="왼손잡이">왼손잡이</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="합계">합계</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td headers="sex" class="gt_row gt_left">남자</td> <td headers="오른손잡이" class="gt_row gt_right">49% (33)</td> <td headers="왼손잡이" class="gt_row gt_right">66% (21)</td> <td headers="합계" class="gt_row gt_right">54% (54)</td></tr> <tr><td headers="sex" class="gt_row gt_left">여자</td> <td headers="오른손잡이" class="gt_row gt_right">51% (35)</td> <td headers="왼손잡이" class="gt_row gt_right">34% (11)</td> <td headers="합계" class="gt_row gt_right">46% (46)</td></tr> <tr><td headers="sex" class="gt_row gt_left">합계</td> <td headers="오른손잡이" class="gt_row gt_right">100% (68)</td> <td headers="왼손잡이" class="gt_row gt_right">100% (32)</td> <td headers="합계" class="gt_row gt_right">100% (100)</td></tr> </tbody> </table> </div> ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% tabyl(sex, handed) %>% adorn_totals(where = c("row", "col"), name = "합계") %>% adorn_percentages(denominator = "col") %>% adorn_pct_formatting(digits = 0) %>% adorn_ns(position = "rear") %>% # 표 외양 gt() %>% tab_header( title = md("**☛ 2변량 교차 범주형 데이터 ☚**"), ) %>% cols_align( align = "right", columns = c(오른손잡이, 왼손잡이, 합계) ) %>% * tab_options( * table.width = pct(70), * heading.background.color = "#1E61B0", # R logo 파란색 * heading.title.font.size = "17px", * column_labels.background.color = "#F7F7F7", # R logo 회색 * column_labels.font.weight = "bold", * stub.background.color = "#ffffff", * stub.font.weight = "bold" * ) ``` ] .panel2-gt_table-auto[ <div id="vejcevtiol" style="padding-left:0px;padding-right:0px;padding-top:10px;padding-bottom:10px;overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #vejcevtiol .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: 70%; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #vejcevtiol .gt_heading { background-color: #1E61B0; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #vejcevtiol .gt_caption { padding-top: 4px; padding-bottom: 4px; } #vejcevtiol .gt_title { color: #FFFFFF; font-size: 17px; font-weight: initial; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #vejcevtiol .gt_subtitle { color: #FFFFFF; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; border-top-color: #FFFFFF; border-top-width: 0; } #vejcevtiol .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #vejcevtiol .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #vejcevtiol .gt_col_heading { color: #333333; background-color: #F7F7F7; font-size: 100%; font-weight: bold; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #vejcevtiol .gt_column_spanner_outer { color: #333333; background-color: #F7F7F7; font-size: 100%; font-weight: bold; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #vejcevtiol .gt_column_spanner_outer:first-child { padding-left: 0; } #vejcevtiol .gt_column_spanner_outer:last-child { padding-right: 0; } #vejcevtiol .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #vejcevtiol .gt_group_heading { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; text-align: left; } #vejcevtiol .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #vejcevtiol .gt_from_md > :first-child { margin-top: 0; } #vejcevtiol .gt_from_md > :last-child { margin-bottom: 0; } #vejcevtiol .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #vejcevtiol .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: bold; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; } #vejcevtiol .gt_stub_row_group { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; vertical-align: top; } #vejcevtiol .gt_row_group_first td { border-top-width: 2px; } #vejcevtiol .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #vejcevtiol .gt_first_summary_row { border-top-style: solid; border-top-color: #D3D3D3; } #vejcevtiol .gt_first_summary_row.thick { border-top-width: 2px; } #vejcevtiol .gt_last_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #vejcevtiol .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #vejcevtiol .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #vejcevtiol .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #vejcevtiol .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #vejcevtiol .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #vejcevtiol .gt_footnote { margin: 0px; font-size: 90%; padding-left: 4px; padding-right: 4px; padding-left: 5px; padding-right: 5px; } #vejcevtiol .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #vejcevtiol .gt_sourcenote { font-size: 90%; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; } #vejcevtiol .gt_left { text-align: left; } #vejcevtiol .gt_center { text-align: center; } #vejcevtiol .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #vejcevtiol .gt_font_normal { font-weight: normal; } #vejcevtiol .gt_font_bold { font-weight: bold; } #vejcevtiol .gt_font_italic { font-style: italic; } #vejcevtiol .gt_super { font-size: 65%; } #vejcevtiol .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 75%; vertical-align: 0.4em; } #vejcevtiol .gt_asterisk { font-size: 100%; vertical-align: 0; } #vejcevtiol .gt_indent_1 { text-indent: 5px; } #vejcevtiol .gt_indent_2 { text-indent: 10px; } #vejcevtiol .gt_indent_3 { text-indent: 15px; } #vejcevtiol .gt_indent_4 { text-indent: 20px; } #vejcevtiol .gt_indent_5 { text-indent: 25px; } </style> <table class="gt_table"> <thead class="gt_header"> <tr> <td colspan="4" class="gt_heading gt_title gt_font_normal gt_bottom_border" style><strong>☛ 2변량 교차 범주형 데이터 ☚</strong></td> </tr> </thead> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1" scope="col" id="sex">sex</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="오른손잡이">오른손잡이</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="왼손잡이">왼손잡이</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="합계">합계</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td headers="sex" class="gt_row gt_left">남자</td> <td headers="오른손잡이" class="gt_row gt_right">39% (25)</td> <td headers="왼손잡이" class="gt_row gt_right">47% (17)</td> <td headers="합계" class="gt_row gt_right">42% (42)</td></tr> <tr><td headers="sex" class="gt_row gt_left">여자</td> <td headers="오른손잡이" class="gt_row gt_right">61% (39)</td> <td headers="왼손잡이" class="gt_row gt_right">53% (19)</td> <td headers="합계" class="gt_row gt_right">58% (58)</td></tr> <tr><td headers="sex" class="gt_row gt_left">합계</td> <td headers="오른손잡이" class="gt_row gt_right">100% (64)</td> <td headers="왼손잡이" class="gt_row gt_right">100% (36)</td> <td headers="합계" class="gt_row gt_right">100% (100)</td></tr> </tbody> </table> </div> ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% tabyl(sex, handed) %>% adorn_totals(where = c("row", "col"), name = "합계") %>% adorn_percentages(denominator = "col") %>% adorn_pct_formatting(digits = 0) %>% adorn_ns(position = "rear") %>% # 표 외양 gt() %>% tab_header( title = md("**☛ 2변량 교차 범주형 데이터 ☚**"), ) %>% cols_align( align = "right", columns = c(오른손잡이, 왼손잡이, 합계) ) %>% tab_options( table.width = pct(70), heading.background.color = "#1E61B0", # R logo 파란색 heading.title.font.size = "17px", column_labels.background.color = "#F7F7F7", # R logo 회색 column_labels.font.weight = "bold", stub.background.color = "#ffffff", stub.font.weight = "bold" ) %>% * tab_spanner(label = "요약 통계량", columns = c(오른손잡이, 왼손잡이)) ``` ] .panel2-gt_table-auto[ <div id="qqkkauwrhy" style="padding-left:0px;padding-right:0px;padding-top:10px;padding-bottom:10px;overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #qqkkauwrhy .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: 70%; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #qqkkauwrhy .gt_heading { background-color: #1E61B0; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #qqkkauwrhy .gt_caption { padding-top: 4px; padding-bottom: 4px; } #qqkkauwrhy .gt_title { color: #FFFFFF; font-size: 17px; font-weight: initial; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #qqkkauwrhy .gt_subtitle { color: #FFFFFF; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; border-top-color: #FFFFFF; border-top-width: 0; } #qqkkauwrhy .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #qqkkauwrhy .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #qqkkauwrhy .gt_col_heading { color: #333333; background-color: #F7F7F7; font-size: 100%; font-weight: bold; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #qqkkauwrhy .gt_column_spanner_outer { color: #333333; background-color: #F7F7F7; font-size: 100%; font-weight: bold; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #qqkkauwrhy .gt_column_spanner_outer:first-child { padding-left: 0; } #qqkkauwrhy .gt_column_spanner_outer:last-child { padding-right: 0; } #qqkkauwrhy .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #qqkkauwrhy .gt_group_heading { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; text-align: left; } #qqkkauwrhy .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #qqkkauwrhy .gt_from_md > :first-child { margin-top: 0; } #qqkkauwrhy .gt_from_md > :last-child { margin-bottom: 0; } #qqkkauwrhy .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #qqkkauwrhy .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: bold; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; } #qqkkauwrhy .gt_stub_row_group { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; vertical-align: top; } #qqkkauwrhy .gt_row_group_first td { border-top-width: 2px; } #qqkkauwrhy .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #qqkkauwrhy .gt_first_summary_row { border-top-style: solid; border-top-color: #D3D3D3; } #qqkkauwrhy .gt_first_summary_row.thick { border-top-width: 2px; } #qqkkauwrhy .gt_last_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #qqkkauwrhy .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #qqkkauwrhy .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #qqkkauwrhy .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #qqkkauwrhy .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #qqkkauwrhy .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #qqkkauwrhy .gt_footnote { margin: 0px; font-size: 90%; padding-left: 4px; padding-right: 4px; padding-left: 5px; padding-right: 5px; } #qqkkauwrhy .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #qqkkauwrhy .gt_sourcenote { font-size: 90%; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; } #qqkkauwrhy .gt_left { text-align: left; } #qqkkauwrhy .gt_center { text-align: center; } #qqkkauwrhy .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #qqkkauwrhy .gt_font_normal { font-weight: normal; } #qqkkauwrhy .gt_font_bold { font-weight: bold; } #qqkkauwrhy .gt_font_italic { font-style: italic; } #qqkkauwrhy .gt_super { font-size: 65%; } #qqkkauwrhy .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 75%; vertical-align: 0.4em; } #qqkkauwrhy .gt_asterisk { font-size: 100%; vertical-align: 0; } #qqkkauwrhy .gt_indent_1 { text-indent: 5px; } #qqkkauwrhy .gt_indent_2 { text-indent: 10px; } #qqkkauwrhy .gt_indent_3 { text-indent: 15px; } #qqkkauwrhy .gt_indent_4 { text-indent: 20px; } #qqkkauwrhy .gt_indent_5 { text-indent: 25px; } </style> <table class="gt_table"> <thead class="gt_header"> <tr> <td colspan="4" class="gt_heading gt_title gt_font_normal gt_bottom_border" style><strong>☛ 2변량 교차 범주형 데이터 ☚</strong></td> </tr> </thead> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="2" colspan="1" scope="col" id="sex">sex</th> <th class="gt_center gt_columns_top_border gt_column_spanner_outer" rowspan="1" colspan="2" scope="colgroup" id="요약 통계량"> <span class="gt_column_spanner">요약 통계량</span> </th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="2" colspan="1" scope="col" id="합계">합계</th> </tr> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="오른손잡이">오른손잡이</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="왼손잡이">왼손잡이</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td headers="sex" class="gt_row gt_left">남자</td> <td headers="오른손잡이" class="gt_row gt_right">46% (30)</td> <td headers="왼손잡이" class="gt_row gt_right">57% (20)</td> <td headers="합계" class="gt_row gt_right">50% (50)</td></tr> <tr><td headers="sex" class="gt_row gt_left">여자</td> <td headers="오른손잡이" class="gt_row gt_right">54% (35)</td> <td headers="왼손잡이" class="gt_row gt_right">43% (15)</td> <td headers="합계" class="gt_row gt_right">50% (50)</td></tr> <tr><td headers="sex" class="gt_row gt_left">합계</td> <td headers="오른손잡이" class="gt_row gt_right">100% (65)</td> <td headers="왼손잡이" class="gt_row gt_right">100% (35)</td> <td headers="합계" class="gt_row gt_right">100% (100)</td></tr> </tbody> </table> </div> ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% tabyl(sex, handed) %>% adorn_totals(where = c("row", "col"), name = "합계") %>% adorn_percentages(denominator = "col") %>% adorn_pct_formatting(digits = 0) %>% adorn_ns(position = "rear") %>% # 표 외양 gt() %>% tab_header( title = md("**☛ 2변량 교차 범주형 데이터 ☚**"), ) %>% cols_align( align = "right", columns = c(오른손잡이, 왼손잡이, 합계) ) %>% tab_options( table.width = pct(70), heading.background.color = "#1E61B0", # R logo 파란색 heading.title.font.size = "17px", column_labels.background.color = "#F7F7F7", # R logo 회색 column_labels.font.weight = "bold", stub.background.color = "#ffffff", stub.font.weight = "bold" ) %>% tab_spanner(label = "요약 통계량", columns = c(오른손잡이, 왼손잡이)) %>% * tab_style( * style = list( * cell_fill(color = "lightblue"), * cell_text( * align = "right", * size = "large", * weight = "bold") * ), * locations = cells_body(rows = sex == "합계", * columns = 합계)) ``` ] .panel2-gt_table-auto[ <div id="xzntiuhcjq" style="padding-left:0px;padding-right:0px;padding-top:10px;padding-bottom:10px;overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #xzntiuhcjq .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: 70%; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #xzntiuhcjq .gt_heading { background-color: #1E61B0; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #xzntiuhcjq .gt_caption { padding-top: 4px; padding-bottom: 4px; } #xzntiuhcjq .gt_title { color: #FFFFFF; font-size: 17px; font-weight: initial; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #xzntiuhcjq .gt_subtitle { color: #FFFFFF; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; border-top-color: #FFFFFF; border-top-width: 0; } #xzntiuhcjq .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #xzntiuhcjq .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #xzntiuhcjq .gt_col_heading { color: #333333; background-color: #F7F7F7; font-size: 100%; font-weight: bold; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #xzntiuhcjq .gt_column_spanner_outer { color: #333333; background-color: #F7F7F7; font-size: 100%; font-weight: bold; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #xzntiuhcjq .gt_column_spanner_outer:first-child { padding-left: 0; } #xzntiuhcjq .gt_column_spanner_outer:last-child { padding-right: 0; } #xzntiuhcjq .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #xzntiuhcjq .gt_group_heading { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; text-align: left; } #xzntiuhcjq .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #xzntiuhcjq .gt_from_md > :first-child { margin-top: 0; } #xzntiuhcjq .gt_from_md > :last-child { margin-bottom: 0; } #xzntiuhcjq .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #xzntiuhcjq .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: bold; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; } #xzntiuhcjq .gt_stub_row_group { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; vertical-align: top; } #xzntiuhcjq .gt_row_group_first td { border-top-width: 2px; } #xzntiuhcjq .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #xzntiuhcjq .gt_first_summary_row { border-top-style: solid; border-top-color: #D3D3D3; } #xzntiuhcjq .gt_first_summary_row.thick { border-top-width: 2px; } #xzntiuhcjq .gt_last_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #xzntiuhcjq .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #xzntiuhcjq .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #xzntiuhcjq .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #xzntiuhcjq .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #xzntiuhcjq .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #xzntiuhcjq .gt_footnote { margin: 0px; font-size: 90%; padding-left: 4px; padding-right: 4px; padding-left: 5px; padding-right: 5px; } #xzntiuhcjq .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #xzntiuhcjq .gt_sourcenote { font-size: 90%; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; } #xzntiuhcjq .gt_left { text-align: left; } #xzntiuhcjq .gt_center { text-align: center; } #xzntiuhcjq .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #xzntiuhcjq .gt_font_normal { font-weight: normal; } #xzntiuhcjq .gt_font_bold { font-weight: bold; } #xzntiuhcjq .gt_font_italic { font-style: italic; } #xzntiuhcjq .gt_super { font-size: 65%; } #xzntiuhcjq .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 75%; vertical-align: 0.4em; } #xzntiuhcjq .gt_asterisk { font-size: 100%; vertical-align: 0; } #xzntiuhcjq .gt_indent_1 { text-indent: 5px; } #xzntiuhcjq .gt_indent_2 { text-indent: 10px; } #xzntiuhcjq .gt_indent_3 { text-indent: 15px; } #xzntiuhcjq .gt_indent_4 { text-indent: 20px; } #xzntiuhcjq .gt_indent_5 { text-indent: 25px; } </style> <table class="gt_table"> <thead class="gt_header"> <tr> <td colspan="4" class="gt_heading gt_title gt_font_normal gt_bottom_border" style><strong>☛ 2변량 교차 범주형 데이터 ☚</strong></td> </tr> </thead> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="2" colspan="1" scope="col" id="sex">sex</th> <th class="gt_center gt_columns_top_border gt_column_spanner_outer" rowspan="1" colspan="2" scope="colgroup" id="요약 통계량"> <span class="gt_column_spanner">요약 통계량</span> </th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="2" colspan="1" scope="col" id="합계">합계</th> </tr> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="오른손잡이">오른손잡이</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="왼손잡이">왼손잡이</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td headers="sex" class="gt_row gt_left">남자</td> <td headers="오른손잡이" class="gt_row gt_right">57% (46)</td> <td headers="왼손잡이" class="gt_row gt_right">68% (13)</td> <td headers="합계" class="gt_row gt_right">59% (59)</td></tr> <tr><td headers="sex" class="gt_row gt_left">여자</td> <td headers="오른손잡이" class="gt_row gt_right">43% (35)</td> <td headers="왼손잡이" class="gt_row gt_right">32% (6)</td> <td headers="합계" class="gt_row gt_right">41% (41)</td></tr> <tr><td headers="sex" class="gt_row gt_left">합계</td> <td headers="오른손잡이" class="gt_row gt_right">100% (81)</td> <td headers="왼손잡이" class="gt_row gt_right">100% (19)</td> <td headers="합계" class="gt_row gt_right" style="background-color: #ADD8E6; font-size: large; text-align: right; font-weight: bold;">100% (100)</td></tr> </tbody> </table> </div> ] --- count: false ##### 데이터 → 요약통계 → 표 .panel1-gt_table-auto[ ```r library(gt); library(janitor) handed_data <- tibble(sex = sample(x = c("남자","여자"), size = 100, replace = TRUE), handed = sample(x = c("왼손잡이", "오른손잡이"), size = 100, replace = T, prob = c(.3,.7))) # 표 handed_data %>% tabyl(sex, handed) %>% adorn_totals(where = c("row", "col"), name = "합계") %>% adorn_percentages(denominator = "col") %>% adorn_pct_formatting(digits = 0) %>% adorn_ns(position = "rear") %>% # 표 외양 gt() %>% tab_header( title = md("**☛ 2변량 교차 범주형 데이터 ☚**"), ) %>% cols_align( align = "right", columns = c(오른손잡이, 왼손잡이, 합계) ) %>% tab_options( table.width = pct(70), heading.background.color = "#1E61B0", # R logo 파란색 heading.title.font.size = "17px", column_labels.background.color = "#F7F7F7", # R logo 회색 column_labels.font.weight = "bold", stub.background.color = "#ffffff", stub.font.weight = "bold" ) %>% tab_spanner(label = "요약 통계량", columns = c(오른손잡이, 왼손잡이)) %>% tab_style( style = list( cell_fill(color = "lightblue"), cell_text( align = "right", size = "large", weight = "bold") ), locations = cells_body(rows = sex == "합계", columns = 합계)) ``` ] .panel2-gt_table-auto[ <div id="grzpzoqxsr" style="padding-left:0px;padding-right:0px;padding-top:10px;padding-bottom:10px;overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #grzpzoqxsr .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: 70%; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #grzpzoqxsr .gt_heading { background-color: #1E61B0; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #grzpzoqxsr .gt_caption { padding-top: 4px; padding-bottom: 4px; } #grzpzoqxsr .gt_title { color: #FFFFFF; font-size: 17px; font-weight: initial; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #grzpzoqxsr .gt_subtitle { color: #FFFFFF; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; border-top-color: #FFFFFF; border-top-width: 0; } #grzpzoqxsr .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #grzpzoqxsr .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #grzpzoqxsr .gt_col_heading { color: #333333; background-color: #F7F7F7; font-size: 100%; font-weight: bold; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #grzpzoqxsr .gt_column_spanner_outer { color: #333333; background-color: #F7F7F7; font-size: 100%; font-weight: bold; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #grzpzoqxsr .gt_column_spanner_outer:first-child { padding-left: 0; } #grzpzoqxsr .gt_column_spanner_outer:last-child { padding-right: 0; } #grzpzoqxsr .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #grzpzoqxsr .gt_group_heading { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; text-align: left; } #grzpzoqxsr .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #grzpzoqxsr .gt_from_md > :first-child { margin-top: 0; } #grzpzoqxsr .gt_from_md > :last-child { margin-bottom: 0; } #grzpzoqxsr .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #grzpzoqxsr .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: bold; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; } #grzpzoqxsr .gt_stub_row_group { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 5px; padding-right: 5px; vertical-align: top; } #grzpzoqxsr .gt_row_group_first td { border-top-width: 2px; } #grzpzoqxsr .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #grzpzoqxsr .gt_first_summary_row { border-top-style: solid; border-top-color: #D3D3D3; } #grzpzoqxsr .gt_first_summary_row.thick { border-top-width: 2px; } #grzpzoqxsr .gt_last_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #grzpzoqxsr .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #grzpzoqxsr .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #grzpzoqxsr .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #grzpzoqxsr .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #grzpzoqxsr .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #grzpzoqxsr .gt_footnote { margin: 0px; font-size: 90%; padding-left: 4px; padding-right: 4px; padding-left: 5px; padding-right: 5px; } #grzpzoqxsr .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #grzpzoqxsr .gt_sourcenote { font-size: 90%; padding-top: 4px; padding-bottom: 4px; padding-left: 5px; padding-right: 5px; } #grzpzoqxsr .gt_left { text-align: left; } #grzpzoqxsr .gt_center { text-align: center; } #grzpzoqxsr .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #grzpzoqxsr .gt_font_normal { font-weight: normal; } #grzpzoqxsr .gt_font_bold { font-weight: bold; } #grzpzoqxsr .gt_font_italic { font-style: italic; } #grzpzoqxsr .gt_super { font-size: 65%; } #grzpzoqxsr .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 75%; vertical-align: 0.4em; } #grzpzoqxsr .gt_asterisk { font-size: 100%; vertical-align: 0; } #grzpzoqxsr .gt_indent_1 { text-indent: 5px; } #grzpzoqxsr .gt_indent_2 { text-indent: 10px; } #grzpzoqxsr .gt_indent_3 { text-indent: 15px; } #grzpzoqxsr .gt_indent_4 { text-indent: 20px; } #grzpzoqxsr .gt_indent_5 { text-indent: 25px; } </style> <table class="gt_table"> <thead class="gt_header"> <tr> <td colspan="4" class="gt_heading gt_title gt_font_normal gt_bottom_border" style><strong>☛ 2변량 교차 범주형 데이터 ☚</strong></td> </tr> </thead> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="2" colspan="1" scope="col" id="sex">sex</th> <th class="gt_center gt_columns_top_border gt_column_spanner_outer" rowspan="1" colspan="2" scope="colgroup" id="요약 통계량"> <span class="gt_column_spanner">요약 통계량</span> </th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="2" colspan="1" scope="col" id="합계">합계</th> </tr> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="오른손잡이">오른손잡이</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" scope="col" id="왼손잡이">왼손잡이</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td headers="sex" class="gt_row gt_left">남자</td> <td headers="오른손잡이" class="gt_row gt_right">49% (33)</td> <td headers="왼손잡이" class="gt_row gt_right">58% (19)</td> <td headers="합계" class="gt_row gt_right">52% (52)</td></tr> <tr><td headers="sex" class="gt_row gt_left">여자</td> <td headers="오른손잡이" class="gt_row gt_right">51% (34)</td> <td headers="왼손잡이" class="gt_row gt_right">42% (14)</td> <td headers="합계" class="gt_row gt_right">48% (48)</td></tr> <tr><td headers="sex" class="gt_row gt_left">합계</td> <td headers="오른손잡이" class="gt_row gt_right">100% (67)</td> <td headers="왼손잡이" class="gt_row gt_right">100% (33)</td> <td headers="합계" class="gt_row gt_right" style="background-color: #ADD8E6; font-size: large; text-align: right; font-weight: bold;">100% (100)</td></tr> </tbody> </table> </div> ] <style> .panel1-gt_table-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-gt_table-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-gt_table-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style>